By Ryan Hamnett, PhD
Numerous biochemical techniques are reliant on the highly selective binding capabilities of antibodies to study proteins of interest in biological pathways and disease states, including western blot, immunohistochemistry (IHC), immunoprecipitation (IP), flow cytometry and ELISA.
An ideal antibody has high sensitivity to detect a target protein even at low expression levels, and high specificity so that it binds only to the protein of interest. Validating the specificity of an antibody is therefore critical to ensure that results are correctly interpreted. The gold standard method for validating antibody specificity is referred to as knockout (KO) validation,1–3 in which a wild-type sample is compared to a sample that has been genetically modified to lack the protein of interest.
Antibodies.com has incorporated knockout validation, using CRISPR/Cas9, as part of its standard antibody validation process. The Knockout Validated seal indicates that an antibody, in addition to being validated in the recommended applications and species, has had its specificity confirmed by knockout validation.
By their nature and immunological function, antibodies bind to their targets with high specificity and affinity. Not all antibodies are exclusive to their intended targets, however, demonstrating cross-reactivity with unrelated proteins, potentially with an even higher affinity than for the protein of interest. This is a significant contributor to the issue of reproducibility within biomedical science.4–7 Not only are laboratories unable to replicate published results with alternative techniques or other antibodies, it also creates a significant economic burden due to wasted time and resources.6,7
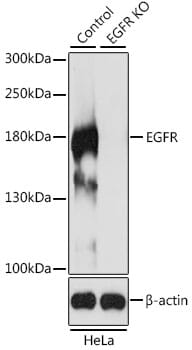
Knockout validation is the first of five pillars of antibody validation established by the independent International Working Group for Antibody Validation.1 Assaying an antibody against KO samples provides confidence that the antibody is binding to the expected protein (Figure 1) and is a key step in addressing the issue of reproducibility.
Figure 1: Western blot analysis of extracts from wild-type (control) and EGFR knockout (KO) HeLa cells using Anti-EGFR antibody (1:500).
The Working Group identified genetic approaches as highly valuable for validation because the link between gene, protein and antibody is so direct.1 KO strategies are also reliable, particularly CRISPR-Cas9, and provide an unambiguous readout (signal vs no signal) that can be assayed with any biochemical technique.
Western blot is one of the most common ways to determine specificity for an antibody. Not only does it indicate specificity by the presence of the target band, it also shows non-specific binding by the presence of other bands at different molecular weights. Three scenarios are depicted in Figure 2 below that could occur when validating an antibody by genetic KO.
Figure 2: Measuring antibody specificity by western blot. A, The ideal situation, in which an antibody shows a single band in the wild-type (WT) condition that is absent in the knockout (KO) condition. Loading controls in the blot at the bottom show that total protein content is consistent between samples. B, Antibody shows non-specific binding to several other proteins, represented by the extra bands, though the specific target band is not present in the KO. Such an antibody may still be useful in some applications. C, Specific band is still present, albeit dimmer, in the KO condition. In this case, the antibody may also be binding to a homologue of similar size. M: marker; WT: wild-type; KO: knockout.
A genetic knockout refers to altering the underlying DNA sequence of a gene to prevent that gene from being transcribed or translated into a functional protein. While this can be performed in any experimental organism, it is most commonly used with cell lines, which can be easily modified with modern molecular biology techniques.
In some instances, a protein is essential to the functioning of the cell and so knocking out its gene results in cell death. In this instance, a knockdown using small interfering or short hairpin RNA (siRNA/shRNA) may be preferred, whereby mRNA is intercepted to prevent translation. This is rarely 100% effective, meaning sufficient protein may be produced to fulfill its essential function and prevent cell death. Knockdowns are also used to validate antibodies,1 but are more prone to off-target effects and so are less precise than knockouts.8–10
Several different methods exist for producing genetic knockout organisms or cells, including homologous recombination, TALENs, and CRISPR-Cas9.
CRISPR-Cas9 is a tool developed in 2012 as a powerful and versatile approach to gene editing,11 for which Jennifer Doudna and Emmanuelle Charpentier were awarded the Nobel Prize in Chemistry in 2020. Co-opting prokaryotic defense mechanisms, CRISPR-Cas9 consists of a ‘guide’ RNA (gRNA) that directs the Cas9 endonuclease enzyme to a specific site in the genome, where Cas9 introduces a double-strand break. When repairing this break, insertions or deletions (indels) can occur that cause a frame-shift mutation in the gene, resulting in a genetic knockout.
CRISPR-Cas9 is easily implemented in an organism of choice and can be directed to any gene by simply altering the gRNA sequence. Thanks to its greater flexibility, efficiency, and specificity compared to other gene editing technologies,12 CRISPR-Cas9 has become the method of choice for genetic engineering, including knockout validation.
In addition to KO validation, we perform validations on species reactivity, applications, and protein arrays.
The International Working Group for Antibody Validation also identified four other pillars of antibody validation following knockout validation:1
Browse our popular KO validated antibodies by research area below.
| Antibody | Applications | Reactivity |
|---|---|---|
| Anti-AIF Antibody | WB, IHC, IF, IP | Human, Mouse, Rat |
| Anti-CDK6 Antibody | WB, IHC, IF | Human, Mouse, Rat |
| Anti-XIAP Antibody | WB, IHC, IF | Human, Mouse, Rat |
| Antibody | Applications | Reactivity |
|---|---|---|
| Anti-hnRNP A2B1 Antibody | WB, IHC, IF | Human, Mouse, Rat |
| Anti-SUZ12 Antibody | WB, IHC | Human, Mouse |
| Anti-WSTF Antibody | WB | Human, Mouse, Rat |
| Antibody | Applications | Reactivity |
|---|---|---|
| Anti-CAPG Antibody | WB, IHC | Human, Mouse, Rat |
| Anti-PD1 Antibody | WB | Human, Mouse, Rat |
| Anti-Transferrin Receptor Antibody | WB, IHC | Human, Mouse, Rat |
| Antibody | Applications | Reactivity |
|---|---|---|
| Anti-IDE Antibody | WB, IHC, IF | Human, Mouse, Rat |
| Anti-Glutamine Synthetase Antibody | WB, IHC, IF | Human, Mouse, Rat |
| Anti-Vimentin Antibody [VI-10] | IP, WB, IHC, ICC | Human, Mouse, Rat |
| Antibody | Applications | Reactivity |
|---|---|---|
| Anti-EGFR Antibody | WB, IHC, ICC/IF, IP | Human, Mouse, Rat |
| Anti-N-Cadherin Antibody [ARC0371] | WB, IHC, ICC/IF | Human, Mouse, Rat |
| Anti-Smad4 Antibody [ARC5009-06] | WB, IHC, IP, ChIP | Human, Mouse, Rat |