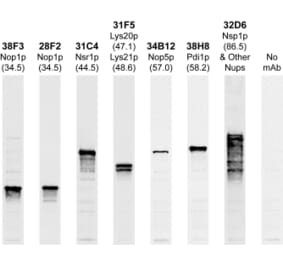
Unconjugated
The data presented here pertain to the research article entitled "Proteome Analysis of Human Embryonic Stem Cell Organelles" (Shekariet al., 2017 [1]). In the present article we endeavour to locate new proteins and pathways in human embryonic stem cells (hESCs) by mass spectrometry and bioinformatics analysis. We have analyzed total and mitochondrial proteins extracted from three biological replicates of the hESC H9 cell line according to mass spectrometry proteomics and bioinformatics investigations.
An intronic GGGGCC expansion in C9orf72 is the most common known cause of both frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS). The repeat expansion leads to the generation of sense and antisense repeat RNA aggregates and dipeptide repeat (DPR) proteins, generated by repeat-associated non-ATG translation. The arginine-rich DPR proteins poly(glycine-arginine or GR) and poly(proline-arginine or PR) are potently neurotoxic and can localise to the nucleolus when expressed in cells, resulting in enlarged nucleoli with disrupted functionality. Furthermore, GGGGCC repeat RNA can bind nucleolar proteins in vitro. However, the relevance of nucleolar stress is unclear, as the arginine-rich DPR proteins do not localise to the nucleolus in C9orf72-associated FTLD/ALS (C9FTLD/ALS) patient brain. We measured nucleolar size in C9FTLD frontal cortex neurons using a three-dimensional, volumetric approach. Intriguingly, we found that C9FTLD brain exhibited bidirectional nucleolar stress. C9FTLD neuronal nucleoli were significantly smaller than control neuronal nucleoli. However, within C9FTLD brains, neurons containing poly(GR) inclusions had significantly larger nucleolar volumes than neurons without poly(GR) inclusions. In addition, expression of poly(GR) in adult Drosophila neurons led to significantly enlarged nucleoli. A small but significant increase in nucleolar volume was also observed in C9FTLD frontal cortex neurons containing GGGGCC repeat-containing RNA foci. These data show that nucleolar abnormalities are a consistent feature of C9FTLD brain, but that diverse pathomechanisms are at play, involving both DPR protein and repeat RNA toxicity.
Cajal bodies (CBs) are important compartments containing accumulated proteins that preferentially regulate RNA-related nuclear events, including splicing. Here, we studied the nuclear distribution pattern of CBs in neurogenesis. In adult brains, coilin was present at a high density, but CB formation was absent in the nuclei of the choroid plexus of the lateral ventricles. Cells of the adult hippocampus were characterized by a crescent-like morphology of coilin protein. We additionally observed a 70 kDa splice variant of coilin in adult mouse brains, which was different to embryonic brains and mouse pluripotent embryonic stem cells (mESCs), characterized by the 80 kDa standard variant of coilin. Here, we also showed that depletion of coilin is induced during neural differentiation and HDAC1 deficiency in mESCs caused coilin accumulation inside the fibrillarin-positive region of the nucleoli. A similar distribution pattern was observed in adult brain hippocampi, characterized by lower levels of both coilin and HDAC1. In summary, we observed that neural differentiation and HDAC1 deficiency lead to coilin depletion and coilin accumulation in body-like structures inside the nucleoli.
Signalling by TGFβ superfamily factors plays an important role in tissue growth and cell proliferation. In Drosophila, the activity of the TGFβ/Activin signalling branch has been linked to the regulation of cell growth and proliferation, but the cellular and molecular basis for these functions are not fully understood. In this study, we show that both the RII receptor Punt (Put) and the R-Smad Smad2 are strongly required for cell and tissue growth. Knocking down the expression of Put or Smad2 in salivary glands causes alterations in nucleolar structure and functions. Cells with decreased TGFβ/Activin signalling accumulate intermediate pre-rRNA transcripts containing internal transcribed spacer 1 regions accompanied by the nucleolar retention of ribosomal proteins. Thus, our results show that TGFβ/Activin signalling is required for ribosomal biogenesis, a key aspect of cellular growth control. Importantly, overexpression of Put enhanced cell growth induced by Drosophila Myc, a well-characterized inducer of nucleolar hypertrophy and ribosome biogenesis.
Digestive organ expansion factor (Def) is a nucleolar protein that plays dual functions: it serves as a component of the ribosomal small subunit processome for the biogenesis of ribosomes and also mediates p53 degradation through the cysteine proteinase calpain-3 (CAPN3). However, nothing is known about the exact relationship between Def and CAPN3 or the regulation of the Def function. In this report, we show that CAPN3 degrades p53 and its mutant proteins p53A138V, p53M237I, p53R248W, and p53R273P but not the p53R175H mutant protein. Importantly, we show that Def directly interacts with CAPN3 in the nucleoli and determines the nucleolar localisation of CAPN3, which is a prerequisite for the degradation of p53 in the nucleolus. Furthermore, we find that Def is modified by phosphorylation at five serine residues: S50, S58, S62, S87, and S92. We further show that simultaneous phosphorylations at S87 and S92 facilitate the nucleolar localisation of Capn3 that is not only essential for the degradation of p53 but is also important for regulating cell cycle progression. Hence, we propose that the Def-CAPN3 pathway serves as a nucleolar checkpoint for cell proliferation by selective inactivation of cell cycle-related substrates during organogenesis.
We have used polysome profiling coupled to microarray analysis to examine the translatome of a panel of peripheral blood (PB) B cells isolated from 34 chronic lymphocytic leukaemia (CLL) patients. We have identified a 'ribosome-related' signature in CLL patients with mRNAs encoding for ribosomal proteins and factors that modify ribosomal RNA, e.g. DKC1 (which encodes dyskerin, a pseudouridine synthase), showing reduced polysomal association and decreased expression of the corresponding proteins. Our data suggest a general impact of dyskerin dysregulation on the translational apparatus in CLL and importantly patients with low dyskerin levels have a significantly shorter period of overall survival following treatment. Thus, translational dysregulation of dyskerin could constitute a mechanism by which the CLL PB B cells acquire an aggressive phenotype and thus have a major role in oncogenesis.
There is growing evidence that defective DNA repair in neurons with accumulation of DNA lesions and loss of genome integrity underlies aging and many neurodegenerative disorders. An important challenge is to understand how neurons can tolerate the accumulation of persistent DNA lesions without triggering the apoptotic pathway. Here we study the impact of the accumulation of unrepaired DNA on the chromatin architecture, kinetics of the DNA damage response and transcriptional activity in rat sensory ganglion neurons exposed to 1-to-3 doses of ionizing radiation (IR). In particular, we have characterized the structural, molecular and transcriptional compartmentalization of unrepaired DNA in persistent DNA damaged foci (PDDF). IR induced the formation of numerous transient foci, which repaired DNA within the 24 h post-IR, and a 1-to-3 PDDF. The latter concentrate DNA damage signaling and repair factors, including γH2AX, pATM, WRAP53 and 53BP1. The number and size of PDDF was dependent on the doses of IR administered. The proportion of neurons carrying PDDF decreased over time of post-IR, indicating that a slow DNA repair occurs in some foci. The fine structure of PDDF consisted of a loose network of unfolded 30 nm chromatin fiber intermediates, which may provide a structural scaffold accessible for DNA repair factors. Furthermore, the transcription assay demonstrated that PDDF are transcriptionally silent, although transcription occurred in flanking euchromatin. Therefore, the expression of γH2AX can be used as a reliable marker of gene silencing in DNA damaged neurons. Moreover, PDDF were located in repressive nuclear environments, preferentially in the perinucleolar domain where they were frequently associated with Cajal bodies or heterochromatin clumps forming a structural triad. We propose that the sequestration of unrepaired DNA in discrete PDDF and the transcriptional silencing can be essential to preserve genome stability and prevent the synthesis of aberrant mRNA and protein products encoded by damaged genes.
The centromere-specific histone CENP-A is the key epigenetic determinant of centromere identity. Whereas most histones are removed from mature sperm, CENP-A is retained to mark paternal centromeres. In Drosophila males we show that the centromere assembly factors CAL1 and CENP-C are required for meiotic chromosome segregation, CENP-A assembly and maintenance on sperm, as well as fertility. In meiosis, CENP-A accumulates with CAL1 in nucleoli. Furthermore, we show that CENP-C normally limits the release of CAL1 and CENP-A from nucleoli for proper centromere assembly in meiotic prophase I. Finally, we show that RNA polymerase I transcription is required for efficient CENP-A assembly in meiosis, as well as centromere tethering to nucleoli.
The Microprocessor complex (DGCR8/Drosha) is required for microRNA (miRNA) biogenesis but also binds and regulates the stability of several types of cellular RNAs. Of particular interest, DGCR8 controls the stability of mature small nucleolar RNA (snoRNA) transcripts independently of Drosha, suggesting the existence of alternative DGCR8 complex(es) with other nucleases to process a variety of cellular RNAs. Here, we found that DGCR8 copurifies with subunits of the nuclear exosome, preferentially associating with its hRRP6-containing nucleolar form. Importantly, we demonstrate that DGCR8 is essential for the recruitment of the exosome to snoRNAs and to human telomerase RNA. In addition, we show that the DGCR8/exosome complex controls the stability of the human telomerase RNA component (hTR/TERC). Altogether, these data suggest that DGCR8 acts as an adaptor to recruit the exosome complex to structured RNAs and induce their degradation.
Actin filaments assemble inside the nucleus in response to multiple cellular perturbations, including heat shock, protein misfolding, integrin engagement, and serum stimulation. We find that DNA damage also generates nuclear actin filaments-detectable by phalloidin and live-cell actin probes-with three characteristic morphologies: (i) long, nucleoplasmic filaments; (ii) short, nucleolus-associated filaments; and (iii) dense, nucleoplasmic clusters. This DNA damage-induced nuclear actin assembly requires two biologically and physically linked nucleation factors: Formin-2 and Spire-1/Spire-2. Formin-2 accumulates in the nucleus after DNA damage, and depletion of either Formin-2 or actin's nuclear import factor, importin-9, increases the number of DNA double-strand breaks (DSBs), linking nuclear actin filaments to efficient DSB clearance. Nuclear actin filaments are also required for nuclear oxidation induced by acute genotoxic stress. Our results reveal a previously unknown role for nuclear actin filaments in DNA repair and identify the molecular mechanisms creating these nuclear filaments.
African horse sickness is a serious equid disease caused by the orbivirus African horse sickness virus (AHSV). The virus has ten double-stranded RNA genome segments encoding seven structural and three non-structural proteins. Recently, an additional protein was predicted to be encoded by genome segment 9 (Seg-9), which also encodes VP6, of most orbiviruses. This has since been confirmed in bluetongue virus and Great Island virus, and the non-structural protein was named NS4. In this study, in silico analysis of AHSV Seg-9 sequences revealed the existence of two main types of AHSV NS4, designated NS4-I and NS4-II, with different lengths and amino acid sequences. The AHSV NS4 coding sequences were in the +1 reading frame relative to that of VP6. Both types of AHSV NS4 were expressed in cultured mammalian cells, with sizes close to the predicted 17-20 kDa. Fluorescence microscopy of these cells revealed a dual cytoplasmic and nuclear, but not nucleolar, distribution that was very similar for NS4-I and NS4-II. Immunohistochemistry on heart, spleen, and lung tissues from AHSV-infected horses showed that NS4 occurs in microvascular endothelial cells and mononuclear phagocytes in all of these tissues, localising to the both the cytoplasm and the nucleus. Interestingly, NS4 was also detected in stellate-shaped dendritic macrophage-like cells with long cytoplasmic processes in the red pulp of the spleen. Finally, nucleic acid protection assays using bacterially expressed recombinant AHSV NS4 showed that both types of AHSV NS4 bind dsDNA, but not dsRNA. Further studies will be required to determine the exact function of AHSV NS4 during viral replication.
The gain-of-function mutant p53 (mtp53) transcriptome has been studied, but, to date, no detailed analysis of the mtp53-associated proteome has been described. We coupled cell fractionation with stable isotope labeling with amino acids in cell culture (SILAC) and inducible knockdown of endogenous mtp53 to determine the mtp53-driven proteome. Our fractionation data highlight the underappreciated biology that missense mtp53 proteins R273H, R280K, and L194F are tightly associated with chromatin. Using SILAC coupled to tandem MS, we identified that R273H mtp53 expression in MDA-MB-468 breast cancer cells up- and down-regulated multiple proteins and metabolic pathways. Here we provide the data set obtained from sequencing 73,154 peptide pairs that then corresponded to 3,010 proteins detected under reciprocal labeling conditions. Importantly, the high impact regulated targets included the previously identified transcriptionally regulated mevalonate pathway proteins but also identified two new levels of mtp53 protein regulation for nontranscriptional targets. Interestingly, mtp53 depletion profoundly influenced poly(ADP ribose) polymerase 1 (PARP1) localization, with increased cytoplasmic and decreased chromatin-associated protein. An enzymatic PARP shift occurred with high mtp53 expression, resulting in increased poly-ADP-ribosylated proteins in the nucleus. Mtp53 increased the level of proliferating cell nuclear antigen (PCNA) and minichromosome maintenance 4 (MCM4) proteins without changing the amount of pcna and mcm4 transcripts. Pathway enrichment analysis ranked the DNA replication pathway above the cholesterol biosynthesis pathway as a R273H mtp53 activated proteomic target. Knowledge of the proteome diversity driven by mtp53 suggests that DNA replication and repair pathways are major targets of mtp53 and highlights consideration of combination chemotherapeutic strategies targeting cholesterol biosynthesis and PARP inhibition.
The Yes-associated protein (YAP) is a potent transcriptional co-activator that functions as a nuclear effector of the Hippo signaling pathway. YAP is oncogenic and its activity is linked to its cellular abundance and nuclear localisation. Activation of the Hippo pathway restricts YAP nuclear entry via its phosphorylation by Lats kinases and consequent cytoplasmic retention bound to 14-3-3 proteins. We examined YAP expression in liver progenitor cells (LPCs) and surprisingly found that transformed LPCs did not show an increase in YAP abundance compared to the non-transformed LPCs from which they were derived. We then sought to ascertain whether nuclear YAP was more abundant in transformed LPCs. We used an antibody that we confirmed was specific for YAP by immunoblotting to determine YAP's sub-cellular localisation by immunofluorescence. This antibody showed diffuse staining for YAP within the cytosol and nuclei, but, noticeably, it showed intense staining of the nucleoli of LPCs. This staining was non-specific, as shRNA treatment of cells abolished YAP expression to undetectable levels by Western blot yet the nucleolar staining remained. Similar spurious YAP nucleolar staining was also seen in mouse embryonic fibroblasts and mouse liver tissue, indicating that this antibody is unsuitable for immunological applications to determine YAP sub-cellular localisation in mouse cells or tissues. Interestingly nucleolar staining was not evident in D645 cells suggesting the antibody may be suitable for use in human cells. Given the large body of published work on YAP in recent years, many of which utilise this antibody, this study raises concerns regarding its use for determining sub-cellular localisation. From a broader perspective, it serves as a timely reminder of the need to perform appropriate controls to ensure the validity of published data.
Hoyeraal-Hreidarsson syndrome (HHS) is a severe form of Dyskeratosis congenita characterized by developmental defects, bone marrow failure and immunodeficiency and has been associated with telomere dysfunction. Recently, mutations in Regulator of Telomere ELongation helicase 1 (RTEL1), a helicase first identified in Mus musculus as being responsible for the maintenance of long telomeres, have been identified in several HHS patients. Here we show that RTEL1 is required for the export and the correct cytoplasmic trafficking of the small nuclear (sn) RNA pre-U2, a component of the major spliceosome complex. RTEL1-HHS cells show abnormal subcellular partitioning of pre-U2, defects in the recycling of ribonucleotide proteins (RNP) in the cytoplasm and splicing defects. While most of these phenotypes can be suppressed by re-expressing the wild-type protein in RTEL1-HHS cells, expression of RTEL1 mutated variants in immortalized cells provokes cytoplasmic mislocalizations of pre-U2 and other RNP components, as well as splicing defects, thus phenocopying RTEL1-HHS cellular defects. Strikingly, expression of a cytoplasmic form of RTEL1 is sufficient to correct RNP mislocalizations both in RTEL1-HHS cells and in cells expressing nuclear mutated forms of RTEL1. This work unravels completely unanticipated roles for RTEL1 in RNP trafficking and strongly suggests that defects in RNP biogenesis pathways contribute to the pathology of HHS.
Both apoptosis ("self-killing") and autophagy ("self-eating") are evolutionarily conserved processes, and their crosstalk influences anticancer drug sensitivity and cell death. However, the underlying mechanism remains unclear. Here, we demonstrated that HMGB1 (high mobility group box 1), normally a nuclear protein, is a crucial regulator of TNFSF10/TRAIL (tumor necrosis factor [ligand] superfamily, member 10)-induced cancer cell death. Activation of PARP1 (poly [ADP-ribose] polymerase 1) was required for TNFSF10-induced ADP-ribosylation of HMGB1 in cancer cells. Moreover, pharmacological inhibition of PARP1 activity or knockdown of PARP1 gene expression significantly inhibited TNFSF10-induced HMGB1 cytoplasmic translocation and subsequent HMGB1-BECN1 complex formation. Furthermore, suppression of the PARP1-HMGB1 pathway diminished autophagy, increased apoptosis, and enhanced the anticancer activity of TNFSF10 in vitro and in a subcutaneous tumor model. These results indicate that PARP1 acts as a prominent upstream regulator of HMGB1-mediated autophagy and maintains a homeostatic balance between apoptosis and autophagy, which provides new insight into the mechanism of TNFSF10 resistance.
Aberrant elevation of JARID1B and histone H3 lysine 4 trimethylation (H3K4me3) is frequently observed in many diseases including prostate cancer (PCa), yet the mechanisms on the regulation of JARID1B and H3K4me3 through epigenetic alterations still remain poorly understood. Here we report that Skp2 modulates JARID1B and H3K4me3 levels in vitro in cultured cells and in vivo in mouse models. We demonstrated that Skp2 inactivation decreased H3K4me3 levels, along with a reduction of cell growth, cell migration and malignant transformation of Pten/Trp53 double null MEFs, and further restrained prostate tumorigenesis of Pten/Trp53 mutant mice. Mechanistically, Skp2 decreased the K63-linked ubiquitination of JARID1B by E3 ubiquitin ligase TRAF6, thus decreasing JARID1B demethylase activity and in turn increasing H3K4me3. In agreement, Skp2 deficiency resulted in an increase of JARID1B ubiquitination and in turn a reduction of H3K4me3, and induced senescence through JARID1B accumulation in nucleoli of PCa cells and prostate tumors of mice. Furthermore, we showed that the elevations of Skp2 and H3K4me3 contributed to castration-resistant prostate cancer (CRPC) in mice, and were positively correlated in human PCa specimens. Taken together, our findings reveal a novel network of SKP2-JARID1B, and targeting SKP2 and JARID1B may be a potential strategy for PCa control.
Chromosome breakage elicits transient silencing of ribosomal RNA synthesis, but the mechanisms involved remained elusive. Here we discover an in trans signalling mechanism that triggers pan-nuclear silencing of rRNA transcription in response to DNA damage. This is associated with transient recruitment of the Nijmegen breakage syndrome protein 1 (NBS1), a central regulator of DNA damage responses, into the nucleoli. We further identify TCOF1 (also known as Treacle), a nucleolar factor implicated in ribosome biogenesis and mutated in Treacher Collins syndrome, as an interaction partner of NBS1, and demonstrate that NBS1 translocation and accumulation in the nucleoli is Treacle dependent. Finally, we provide evidence that Treacle-mediated NBS1 recruitment into the nucleoli regulates rRNA silencing in trans in the presence of distant chromosome breaks.
A major constituent of the nuclear basket region of the nuclear pore complex (NPC), nucleoporin Tpr, plays roles in regulating multiple important processes. We have previously established that Tpr is phosphorylated in both a MAP-kinase-dependent and MAP-kinase-independent manner, and that Tpr acts as both a substrate and as a scaffold for ERK2 (also known as MAPK1). Here, we report the identification of S2059 and S2094 as the major novel ERK-independent phosphorylation sites and T1677, S2020, S2023 and S2034 as additional ERK-independent phosphorylation sites found in the Tpr protein in vivo. Our results suggest that protein kinase A phosphorylates the S2094 residue and that the site is hyperphosphorylated during mitosis. Furthermore, we find that Tpr is phosphorylated at the S2059 residue by CDK1 and the phosphorylated form distinctly localizes with chromatin during telophase. Abrogation of S2059 phosphorylation abolishes the interaction of Tpr with Mad1, thus compromising the localization of both Mad1 and Mad2 proteins, resulting in cell cycle defects. The identification of novel phosphorylation sites on Tpr and the observations presented in this study allow better understanding of Tpr functions.
Cajal bodies are important nuclear structures containing proteins that preferentially regulate RNA-related metabolism. We investigated the cell-type specific nuclear distribution of Cajal bodies and the level of coilin, a protein of Cajal bodies, in non-irradiated and irradiated human tumor cell lines and embryonic stem (ES) cells. Cajal bodies were localized in different nuclear compartments, including DAPI-poor regions, in the proximity of chromocenters, and adjacent to nucleoli. The number of Cajal bodies per nucleus was cell cycle-dependent, with higher numbers occurring during G2 phase. Human ES cells contained a high coilin level in the nucleoplasm, but coilin-positive Cajal bodies were also identified in nuclei of mouse and human ES cells. Coilin, but not SMN, recognized UVA-induced DNA lesions, which was cell cycle-independent. Treatment with γ-radiation reduced the localized movement of Cajal bodies in many cell types and GFP-coilin fluorescence recovery after photobleaching was very fast in nucleoplasm in comparison with GFP-coilin recovery in DNA lesions. By contrast, nucleolus-localized coilin displayed very slow fluorescence recovery after photobleaching, which indicates very slow rates of protein diffusion, especially in nucleoli of mouse ES cells.
EGR1 is an immediate early gene with a wide range of activities as transcription factor, spanning from regulation of cell growth to differentiation. Numerous studies show that EGR1 either promotes the proliferation of stimulated cells or suppresses the tumorigenic growth of transformed cells. Upon interaction with ARF, EGR1 is sumoylated and acquires the ability to bind to specific targets such as PTEN and in turn to regulate cell growth. ARF is mainly localized to the periphery of nucleolus where is able to negatively regulate ribosome biogenesis. Since EGR1 colocalizes with ARF under IGF-1 stimulation we asked the question of whether EGR1 also relocate to the nucleolus to interact with ARF. Here we show that EGR1 colocalizes with nucleolar markers such as fibrillarin and B23 in the presence of ARF. Western analysis of nucleolar extracts from HeLa cells was used to confirm the presence of EGR1 in the nucleolus mainly as the 100 kDa sumoylated form. We also show that the level of the ribosomal RNA precursor 47S is inversely correlated to the level of EGR1 transcripts. The EGR1 iseffective to regulate the synthesis of the 47S rRNA precursor. Then we demonstrated that EGR1 binds to the Upstream Binding Factor (UBF) leading us to hypothesize that the regulating activity of EGR1 is mediated by its interaction within the transcriptional complex of RNA polymerase I. These results confirm the presence of EGR1 in the nucleolus and point to a role for EGR1 in the control of nucleolar metabolism.
We previously identified a Drosophila maternal effect-lethal mutant named 'no poles' (nopo). Embryos from nopo females undergo mitotic arrest with barrel-shaped, acentrosomal spindles during the rapid cycles of syncytial embryogenesis because of activation of a Chk2-mediated DNA checkpoint. NOPO is the Drosophila homolog of human TNF receptor associated factor (TRAF)-interacting protein (TRIP), which has been implicated in TNF signaling. NOPO and TRIP contain RING domains closely resembling those of known E3 ubiquitin ligases. We herein sought to elucidate the mechanism by which TRIP/NOPO promotes genomic stability by performing a yeast two-hybrid screen to identify potential substrates/interactors. We identified members of the Y-family of DNA polymerases that facilitate replicative bypass of damaged DNA (translesion synthesis) as TRIP interactors. We show that TRIP and NOPO co-immunoprecipitate with human and Drosophila Polη, respectively, from cultured cells. We generated a null mutation in Drosophila Polη (dPolη) and found that dPolη-derived embryos have increased sensitivity to ultraviolet irradiation and exhibit nopo-like mitotic spindle defects. dPolη and nopo interact genetically in that overexpression of dPolη in hypomorphic nopo-derived embryos suppresses nopo phenotypes. We observed enhanced ubiquitylation of Polη by TRIP and NOPO E3 ligases in human cells and Drosophila embryos, respectively, and show that TRIP promotes hPolη localization to nuclear foci in human cells. We present a model in which TRIP/NOPO ubiquitylates Polη to positively regulate its activity in translesion synthesis.
To maintain growth and division, cells require a large-scale production of rRNAs which occurs in the nucleolus. Recently, we have shown the interaction of nucleolar phosphatidylinositol 4,5-bisphosphate (PIP2) with proteins involved in rRNA transcription and processing, namely RNA polymerase I (Pol I), UBF, and fibrillarin. Here we extend the study by investigating transcription-related localization of PIP2 in regards to transcription and processing complexes of Pol I. To achieve this, we used either physiological inhibition of transcription during mitosis or inhibition by treatment the cells with actinomycin D (AMD) or 5,6-dichloro-1β-d-ribofuranosyl-benzimidazole (DRB). We show that PIP2 is associated with Pol I subunits and UBF in a transcription-independent manner. On the other hand, PIP2/fibrillarin colocalization is dependent on the production of rRNA. These results indicate that PIP2 is required not only during rRNA production and biogenesis, as we have shown before, but also plays a structural role as an anchor for the Pol I pre-initiation complex during the cell cycle. We suggest that throughout mitosis, PIP2 together with UBF is involved in forming and maintaining the core platform of the rDNA helix structure. Thus we introduce PIP2 as a novel component of the NOR complex, which is further engaged in the renewed rRNA synthesis upon exit from mitosis.
Nucleosomes are decorated with numerous post-translational modifications capable of influencing many DNA processes. Here we describe a new class of histone modification, methylation of glutamine, occurring on yeast histone H2A at position 105 (Q105) and human H2A at Q104. We identify Nop1 as the methyltransferase in yeast and demonstrate that fibrillarin is the orthologue enzyme in human cells. Glutamine methylation of H2A is restricted to the nucleolus. Global analysis in yeast, using an H2AQ105me-specific antibody, shows that this modification is exclusively enriched over the 35S ribosomal DNA transcriptional unit. We show that the Q105 residue is part of the binding site for the histone chaperone FACT (facilitator of chromatin transcription) complex. Methylation of Q105 or its substitution to alanine disrupts binding to FACT in vitro. A yeast strain mutated at Q105 shows reduced histone incorporation and increased transcription at the ribosomal DNA locus. These features are phenocopied by mutations in FACT complex components. Together these data identify glutamine methylation of H2A as the first histone epigenetic mark dedicated to a specific RNA polymerase and define its function as a regulator of FACT interaction with nucleosomes.
During Drosophila oogenesis, the endopolyploid nuclei of germ-line nurse cells undergo a dramatic shift in morphology as oogenesis progresses; the easily-visible chromosomes are initially polytenic during the early stages of oogenesis before they transiently condense into a distinct '5-blob' configuration, with subsequent dispersal into a diffuse state. Mutations in many genes, with diverse cellular functions, can affect the ability of nurse cells to fully decondense their chromatin, resulting in a '5-blob arrest' phenotype that is maintained throughout the later stages of oogenesis. However, the mechanisms and significance of nurse-cell (NC) chromatin dispersal remain poorly understood. Here, we report that a screen for modifiers of the 5-blob phenotype in the germ line isolated the spliceosomal gene peanuts, the Drosophila Prp22. We demonstrate that reduction of spliceosomal activity through loss of peanuts promotes decondensation defects in NC nuclei during mid-oogenesis. We also show that the Prp38 spliceosomal protein accumulates in the nucleoplasm of nurse cells with impaired peanuts function, suggesting that spliceosomal recycling is impaired. Finally, we reveal that loss of additional spliceosomal proteins impairs the full decondensation of NC chromatin during later stages of oogenesis, suggesting that individual spliceosomal subcomplexes modulate expression of the distinct subset of genes that are required for correct morphology in endopolyploid nurse cells.
Human cancers over-expressing mdm2, through a T to G variation at a single nucleotide polymorphism at position 309 (mdm2 SNP309), have functionally inactivated p53 that is not effectively degraded. They also have high expression of the alternatively spliced transcript, mdm2-C. Alternatively spliced mdm2 transcripts are expressed in many forms of human cancer and when they are exogenously expressed they transform human cells. However no study to date has detected endogenous MDM2 protein isoforms. Studies with exogenous expression of splice variants have been carried out with mdm2-A and mdm2-B, but the mdm2-C isoform has remained virtually unexplored. We addressed the cellular influence of exogenously expressed MDM2-C, and asked if endogenous MDM2-C protein was present in human cancers. To detect endogenous MDM2-C protein, we created a human MDM2-C antibody to the splice junction epitope of exons four and ten (MDM2 C410) and validated the antibody with in vitro translated full length MDM2 compared to MDM2-C. Interestingly, we discovered that MDM2-C co-migrates with MDM2-FL at approximately 98 kDa. Using the validated C410 antibody, we detected high expression of endogenous MDM2-C in human cancer cell lines and human cancer tissues. In the estrogen receptor positive (ER+) mdm2 G/G SNP309 breast cancer cell line, T47D, we observed an increase in endogenous MDM2-C protein with estrogen treatment. MDM2-C localized to the nucleus and the cytoplasm. We examined the biological activity of MDM2-C by exogenously expressing the protein and observed that MDM2-C did not efficiently target p53 for degradation or reduce p53 transcriptional activity. Exogenous expression of MDM2-C in p53-null human cancer cells increased colony formation, indicating p53-independent tumorigenic properties. Our data indicate a role for MDM2-C that does not require the inhibition of p53 for increasing cancer cell proliferation and survival.
The evolutionarily conserved JAK/STAT pathway plays important roles in development and disease processes in humans. Although the signaling process has been well established, we know relatively little about what the relevant target genes are that mediate JAK/STAT activation during development. Here, we have used genome-wide microarrays to identify JAK/STAT targets in the optic lobes of the Drosophila brain and identified 47 genes that are positively regulated by JAK/STAT. About two-thirds of the genes encode proteins that have orthologs in humans. The STAT targets in the optic lobe appear to be different from the targets identified in other tissues, suggesting that JAK/STAT signaling may regulate different target genes in a tissue-specific manner. Functional analysis of Nop56, a cell-autonomous STAT target, revealed an essential role for this gene in the growth and proliferation of neuroepithelial stem cells in the optic lobe and an inhibitory role in lamina neurogenesis.
Duplicated ribosomal protein (Rp) gene families often encode highly similar or identical proteins with redundant or unique roles. Eukaryotic-specific paralogues RpL22e and RpL22e-like-PA are structurally divergent within the N terminus and differentially expressed, suggesting tissue-specific functions. We previously identified RpL22e-like-PA as a testis Rp. Strikingly, RpL22e is detected in immunoblots at its expected molecular mass (m) of 33 kD and at increasing m of ~43-55 kD, suggesting RpL22e post-translational modification (PTM). Numerous PTMs, including N-terminal SUMOylation, are predicted computationally. Based on S2 cell co-immunoprecipitations, bacterial-based SUMOylation assays and in vivo germline-specific RNAi depletion of SUMO, we conclude that RpL22e is a SUMO substrate. Testis-specific PTMs are evident, including a phosphorylated version of SUMOylated RpL22e identified by in vitro phosphatase experiments. In ribosomal profiles from S2 cells, only unconjugated RpL22e co-sediments with active ribosomes, supporting an extra-translational role for SUMOylated RpL22e. Ectopic expression of an RpL22e N-terminal deletion (lacking SUMO motifs) shows that truncated RpL22e co-sediments with polysomes, implying that RpL22e SUMOylation is dispensable for ribosome biogenesis and function. In mitotic germ cells, both paralogues localize within the cytoplasm and nucleolus. However, within meiotic cells, phase contrast microscopy and co-immunohistochemical analysis with nucleolar markers nucleostemin1 and fibrillarin reveals diffuse nucleoplasmic, but not nucleolar RpL22e localization that transitions to a punctate pattern as meiotic cells mature, suggesting an RpL22e role outside of translation. Germline-specific knockdown of SUMO shows that RpL22e nucleoplasmic distribution is sensitive to SUMO levels, as immunostaining becomes more dispersed. Overall, these data suggest distinct male germline roles for RpL22e and RpL22e-like-PA.
Pseudouridine is the most abundant modified nucleotide in ribosomal RNA throughout eukaryotes and archaea but its role is not known. Here we produced mouse embryonic fibroblast cells expressing only catalytically inactive dyskerin, the pseudouridine synthase that converts uridine to pseudouridine in ribosomal RNA. The mutant dyskerin protein, D125A, was extremely unstable but cells were able to divide and grow very slowly. Abnormalities in ribosome RNA synthesis were apparent but mature cytoplasmic RNAs lacking pseudouridine were produced and were very unstable. We conclude that pseudouridine is required to stabilize the secondary structure of ribosomal RNA that is essential for its function.
Sepsis is caused by an overwhelming immune response to bacterial infection. The discovery of high mobility group box 1 (HMGB1) as a late mediator of lethal sepsis has prompted investigation into the development of new therapeutics which specifically target this protein. Here, we show that chloroquine, an anti-malarial drug, prevents lethality in mice with established endotoxemia or sepsis. This effect is still observed even if administration of chloroquine is delayed. The protective effects of chloroquine were mediated through inhibition of HMGB1 release in macrophages, monocytes, and endothelial cells, thereby preventing its cytokine-like activities. As an inhibitor of autophagy, chloroquine specifically inhibited HMGB1-induced Iκ-B degradation and NF-κB activation. These findings define a novel mechanism for the anti-inflammatory effects of chloroquine and also suggest a new potential clinical use for this drug in the setting of sepsis.
Digestive organs originate from the endoderm. Morphogenesis of the digestive system is precisely controlled by multiple factors that dictate the cell fate and behavior so that the specific digestive organs are timely formed in the right place and develop into right size and structure. We showed previously that digestive organ expansion factor (def) is a gene whose expression is enriched in the liver, pancreas and intestine. Loss-of-function of def in the def(hi429) mutant confers hypoplastic digestive organs partly due to alteration of expression of genes related to the p53 pathway. However, the molecular mechanism for the involvement of Def in the organogenesis of digestive organs is still largely unknown. For example, it is not known whether Def regulates specific pathways in a specific organ. To address this question, we generated four independent Tg(fabp10a:def) transgenic fish lines which over-expressed Def specifically in the liver. We characterized Tg-I, one of the transgenic lines, in detail with genetic, molecular and histological approaches. We found that Tg-I restored the liver but not exocrine pancreas and intestine development in the def(hi429) mutant. However, Tg-I adult fish in the wild type (WT) background exhibits reduced liver-to-body ratio and all four transgenic lines conferred abnormal intrahepatic structure. Microarray data analysis showed that certain specific functional pathways were affected in the liver of Tg-I. These results demonstrate that Def functions in a cell autonomous manner during early liver development and aberrant Def protein expression might lead to disruption of the structural integrity of a normal adult liver.
The DEAD-box protein UAP56 (U2AF65-associcated protein) is an RNA helicase that in yeast and metazoa is critically involved in mRNA splicing and export. In Arabidopsis, two adjacent genes code for an identical UAP56 protein, and both genes are expressed. In case one of the genes is inactivated by a T-DNA insertion, wild type transcript level is maintained by the other intact gene. In contrast to other organisms that are severely affected by elevated UAP56 levels, Arabidopsis plants that overexpress UAP56 have wild type appearance. UAP56 localises predominantly to euchromatic regions of Arabidopsis nuclei, and associates with genes transcribed by RNA polymerase II independently from the presence of introns, while it is not detected at non-transcribed loci. Biochemical characterisation revealed that in addition to ssRNA and dsRNA, UAP56 interacts with dsDNA, but not with ssDNA. Moreover, the enzyme displays ATPase activity that is stimulated by RNA and dsDNA and it has ATP-dependent RNA helicase activity unwinding dsRNA, whereas it does not unwind dsDNA. Protein interaction studies showed that UAP56 directly interacts with the mRNA export factors ALY2 and MOS11, suggesting that it is involved in mRNA export from plant cell nuclei.
Actin and nuclear myosin 1c (NM1) cooperate in RNA polymerase I (pol I) transcription. NM1 is also part of a multiprotein assembly, B-WICH, which is involved in transcription. This assembly contains the chromatin remodeling complex WICH with its subunits WSTF and SNF2h. We report here that NM1 binds SNF2h with enhanced affinity upon impairment of the actin-binding function. ChIP analysis revealed that NM1, SNF2h, and actin gene occupancies are cell cycle-dependent and require intact motor function. At the onset of cell division, when transcription is temporarily blocked, B-WICH is disassembled due to WSTF phosphorylation, to be reassembled on the active gene at exit from mitosis. NM1 gene knockdown and motor function inhibition, or stable expression of NM1 mutants that do not interact with actin or chromatin, overall repressed rRNA synthesis by stalling pol I at the gene promoter, led to chromatin alterations by changing the state of H3K9 acetylation at gene promoter, and delayed cell cycle progression. These results suggest a unique structural role for NM1 in which the interaction with SNF2h stabilizes B-WICH at the gene promoter and facilitates recruitment of the HAT PCAF. This leads to a permissive chromatin structure required for transcription activation.
When cell cycle withdrawal accompanies terminal differentiation, biosynthesis and cellular growth are likely to change also. In this study, nucleolus size was monitored during cell fate specification in the Drosophila eye imaginal disc using fibrillarin antibody labeling. Nucleolus size is an indicator of ribosome biogenesis and can correlate with cellular growth rate. Nucleolar size was reduced significantly during cell fate specification and differentiation, predominantly as eye disc cells entered a cell cycle arrest that preceded cell fate specification. This reduction in nucleolus size required Dpp and Hh signaling. A transient enlargement of the nucleolus accompanied cell division in the Second Mitotic Wave. Nucleoli continued to diminish in postmitotic cells following fate specification. These results suggest that cellular growth is regulated early in the transition from proliferating progenitor cells to terminal cell fate specification, contemporary with regulation of the cell cycle, and requiring the same extracellular signals.
p53 protein turnover through the ubiquitination pathway is a vital mechanism in the regulation of its transcriptional activity; however, little is known about p53 turnover through proteasome-independent pathway(s). The digestive organ expansion factor (Def) protein is essential for the development of digestive organs. In zebrafish, loss of function of def selectively upregulates the expression of p53 response genes, which raises a question as to what is the relationship between Def and p53. We report here that Def is a nucleolar protein and that loss of function of def leads to the upregulation of p53 protein, which surprisingly accumulates in the nucleoli. Our extensive studies have demonstrated that Def can mediate the degradation of p53 protein and that this process is independent of the proteasome pathway, but dependent on the activity of Calpain3, a cysteine protease. Our findings define a novel nucleolar pathway that regulates the turnover function of p53, which will advance our understanding of p53's role in organogenesis and tumorigenesis.
The abundance of Myc protein must be exquisitely controlled to avoid growth abnormalities caused by too much or too little Myc. An intriguing mode of regulation exists in which Myc protein itself leads to reduction in its abundance. We show here that dMyc binds to the miR-308 locus and increases its expression. Using our gain-of-function approach, we show that an increase in miR-308 causes a destabilization of dMyc mRNA and reduced dMyc protein levels. In vivo knockdown of miR-308 confirmed the regulation of dMyc levels in embryos. This regulatory loop is crucial for maintaining appropriate dMyc levels and normal development. Perturbation of the loop, either by elevated miR-308 or elevated dMyc, caused lethality. Combining elevated levels of both, therefore restoring balance between miR-308 and dMyc levels, resulted in lower apoptotic activity and suppression of lethality. These results reveal a sensitive feedback mechanism that is crucial to prevent the pathologies caused by abnormal levels of dMyc.
The diverse functions of Notch signalling imply that it must elicit context-specific programmes of gene expression. With the aim of investigating how Notch drives cells to differentiate, we have used a genome-wide approach to identify direct Notch targets in Drosophila haemocytes (blood cells), where Notch promotes crystal cell differentiation. Many of the identified Notch-regulated enhancers contain Runx and GATA motifs, and we demonstrate that binding of the Runx protein Lozenge (Lz) is required for enhancers to be competent to respond to Notch. Functional studies of targets, such as klumpfuss (ERG/WT1 family) and pebbled/hindsight (RREB1 homologue), show that Notch acts both to prevent the cells adopting alternate cell fates and to promote morphological characteristics associated with crystal cell differentiation. Inappropriate activity of Klumpfuss perturbs the differentiation programme, resulting in melanotic tumours. Thus, by acting as a master regulator, Lz directs Notch to activate selectively a combination of target genes that correctly locks cells into the differentiation programme.
The organization of the genome within the mammalian nucleus is nonrandom, with physiologic processes often concentrated in specific three-dimensional domains. This organization may be functionally related to gene regulation and, as such, may play a role in normal development and human disease processes. However, the mechanisms that participate in nuclear organization are poorly understood. Here, we present data characterizing localization of the imprinted Kcnq1 alleles. We show that nucleolar association of the paternal allele (1) is stimulated during the differentiation of trophoblast stem cells, (ii) is dependent upon the Kcnq1ot1 noncoding RNA, (3) does not require polycomb repressive complex 2, and (4) is not sufficient to preclude transcription of imprinted genes. Although nucleolar positioning has been proposed as a mechanism to related to gene silencing, we find that silencing and perinucleolar localization through the Kcnq1ot1 noncoding RNA are separable events.
Fundamental to the life and destiny of every cell is the regulation of protein synthesis through ribosome biogenesis, which begins in the nucleolus with the production of ribosomal RNA (rRNA). Nucleolar organization is a highly dynamic and tightly regulated process; the structural factors that direct nucleolar assembly and disassembly are just as important in controlling rRNA synthesis as are the catalytic activities that synthesize rRNA. Here, we report that a signaling enzyme, inositol 1,3,4,5,6-pentakisphosphate 2-kinase (IP5K) is also a structural component in the nucleolus. We demonstrate that IP5K has functionally significant interactions with three proteins that regulate rRNA synthesis: protein kinase CK2, TCOF1 and upstream-binding-factor (UBF). Through molecular modeling and mutagenic studies, we identified an Arg-Lys-Lys tripeptide located on the surface of IP5K that mediates its association with UBF. Nucleolar IP5K spatial dynamics were sensitive to experimental procedures (serum starvation or addition of actinomycin D) that inhibited rRNA production. We show that IP5K makes stoichiometrically sensitive contributions to the architecture of the nucleoli in intact cells, thereby influencing the degree of rRNA synthesis. Our study adds significantly to the biological significance of IP5K; previously, it was the kinase activity of this protein that had attracted attention. Our demonstration that IP5K 'moonlights' as a molecular scaffold offers an unexpected new example of how the biological sophistication of higher organisms can arise from gene products acquiring multiple functions, rather than by an increase in gene number.
Loss of amino groups from adenines in DNA results in the formation of hypoxanthine (Hx) bases with miscoding properties. The primary enzyme in Escherichia coli for DNA repair initiation at deaminated adenine is endonuclease V (endoV), encoded by the nfi gene, which cleaves the second phosphodiester bond 3' of an Hx lesion. Endonuclease V orthologs are widespread in nature and belong to a family of highly conserved proteins. Whereas prokaryotic endoV enzymes are well characterized, the function of the eukaryotic homologs remains obscure. Here we describe the human endoV ortholog and show with bioinformatics and experimental analysis that a large number of transcript variants exist for the human endonuclease V gene (ENDOV), many of which are unlikely to be translated into functional protein. Full-length ENDOV is encoded by 8 evolutionary conserved exons covering the core region of the enzyme, in addition to one or more 3'-exons encoding an unstructured and poorly conserved C-terminus. In contrast to the E. coli enzyme, we find recombinant ENDOV neither to incise nor bind Hx-containing DNA. While both enzymes have strong affinity for several branched DNA substrates, cleavage is observed only with E. coli endoV. We find that ENDOV is localized in the cytoplasm and nucleoli of human cells. As nucleoli harbor the rRNA genes, this may suggest a role for the protein in rRNA gene transactions such as DNA replication or RNA transcription.
In the Caenorhabditis elegans nematode, the oocyte nucleolus disappears prior to fertilization. We have now investigated the re-formation of the nucleolus in the early embryo of this model organism by immunostaining for fibrillarin and DAO-5, a putative NOLC1/Nopp140 homolog involved in ribosome assembly. We find that labeled nucleoli first appear in somatic cells at around the 8-cell stage, at a time when transcription of the embryonic genome begins. Quantitative analysis of radial positioning showed the nucleolus to be localized at the nuclear periphery in a majority of early embryonic nuclei. At the ultrastructural level, the embryonic nucleolus appears to be composed of a relatively homogenous core surrounded by a crescent-shaped granular structure. Prior to embryonic genome activation, fibrillarin and DAO-5 staining is seen in numerous small nucleoplasmic foci. This staining pattern persists in the germline up to the ∼100-cell stage, until the P4 germ cell divides to give rise to the Z2/Z3 primordial germ cells and embryonic transcription is activated in this lineage. In the ncl-1 mutant, which is characterized by increased transcription of rDNA, DAO-5-labeled nucleoli are already present at the 2-cell stage. Our results suggest a link between the activation of transcription and the initial formation of nucleoli in the C. elegans embryo.
CTCF is a ubiquitous epigenetic regulator that has been proposed as a master keeper of chromatin organisation. CTCF-like, or BORIS, is thought to antagonise CTCF and has been found in normal testis, ovary and a large variety of tumour cells. The cellular function of BORIS remains intriguing although it might be involved in developmental reprogramming of gene expression patterns. We here unravel the expression of CTCF and BORIS proteins throughout human epidermis. While CTCF is widely distributed within the nucleus, BORIS is confined to the nucleolus and other euchromatin domains. Nascent RNA experiments in primary keratinocytes revealed that endogenous BORIS is present in active transcription sites. Interestingly, BORIS also localises to interphase centrosomes suggesting a role in the cell cycle. Blocking the cell cycle at S phase or mitosis, or causing DNA damage, produced a striking accumulation of BORIS. Consistently, ectopic expression of wild type or GFP- BORIS provoked a higher rate of S phase cells as well as genomic instability by mitosis failure. Furthermore, down-regulation of endogenous BORIS by specific shRNAs inhibited both RNA transcription and cell cycle progression. The results altogether suggest a role for BORIS in coordinating S phase events with mitosis.
The ability of most cancer cells to grow indefinitely relies on the enzyme telomerase and its recruitment to telomeres. In human cells, recruitment depends on the Cajal body RNA chaperone TCAB1 binding to the RNA subunit of telomerase (hTR) and is also thought to rely on an N-terminal domain of the catalytic subunit, hTERT. We demonstrate that coilin, an essential structural component of Cajal bodies, is required for endogenous telomerase recruitment to telomeres but that overexpression of telomerase can compensate for Cajal body absence. In contrast, recruitment of telomerase was sensitive to levels of TCAB1, and this was not rescued by overexpression of telomerase. Thus, although Cajal bodies are important for recruitment, TCAB1 has an additional role in this process that is independent of these structures. TCAB1 itself localizes to telomeres in a telomerase-dependent but Cajal body-independent manner. We identify a point mutation in hTERT that largely abolishes recruitment yet does not affect association of telomerase with TCAB1, suggesting that this region mediates recruitment by an independent mechanism. Our results demonstrate that telomerase has multiple independent requirements for recruitment to telomeres and that the function of TCAB1 is to directly transport telomerase to telomeres.
Our previous study demonstrated that 45S ribosomal DNA (45S rDNA) clusters were chromosome fragile sites expressed spontaneously in Lolium. In this study, fragile phenotypes of 45S rDNA were observed under aphidicolin (APH) incubation in several plant species. Further actinomycin D (ActD) treatment showed that transcriptional stress might interfere with chromatin packaging, resulting in 45S rDNA fragile expression. These data identified 45S rDNA sites as replication-dependent as well as transcription-dependent fragile sites in plants. In the presence of ActD, a dramatic switch to an open chromatin conformation and accumulated incomplete 5' end of the external transcribed spacer (5'ETS) transcripts were observed, accompanied by decreased DNA methylation, decreased levels of histone H3, and increased histone acetylation and levels of H3K4me2, suggesting that these epigenetic alterations are associated with failure of 45S rDNA condensation. Furthermore, the finding that γ-H2AX was accumulated at 45S rDNA sites following ActD treatment suggested that the DNA damage signaling pathway was associated with the appearance of 45S rDNA fragile phenotypes. Our data provide a link between 45S rDNA transcription and chromatin-packaging defects and open the door for further identifying the molecular mechanism involved.
Histone H1 is an intrinsic component of chromatin, whose important contribution to chromatin structure is well-established in vitro. Little is known, however, about its functional roles in vivo. Here, we have addressed this question in Drosophila, a model system offering many advantages since it contains a single dH1 variant. For this purpose, RNAi was used to efficiently deplete dH1 in flies. Expression-profiling shows that dH1 depletion affects expression of a relatively small number of genes in a regional manner. Furthermore, depletion up-regulates inactive genes, preferentially those located in heterochromatin, while active euchromatic genes are down-regulated, suggesting that the contribution of dH1 to transcription regulation is mainly structural, organizing chromatin for proper gene-expression regulation. Up-regulated genes are remarkably enriched in transposons. In particular, R1/R2 retrotransposons, which specifically integrate in the rDNA locus, are strongly up-regulated. Actually, depletion increases expression of transposon-inserted rDNA copies, resulting in synthesis of aberrant rRNAs and enlarged nucleolus. Concomitantly, dH1-depleted cells accumulate extra-chromosomal rDNA, show increased γH2Av content, stop proliferation and activate apoptosis, indicating that depletion causes genome instability and affects proliferation. Finally, the contributions to maintenance of genome integrity and cell proliferation appear conserved in human hH1s, as their expression rescues proliferation of dH1-depleted cells.
Protein syntheses mediated by cellular and viral internal ribosome entry sites (IRESs) are believed to have many features in common. Distinct mechanisms for ribosome recruitment and preinitiation complex assembly between the two processes have not been identified thus far. Here we show that the methylation status of rRNA differentially influenced the mechanism of 80S complex formation on IRES elements from the cellular sodium-coupled neutral amino acid transporter 2 (SNAT2) versus the hepatitis C virus mRNA. Translation initiation involves the assembly of the 48S preinitiation complex, followed by joining of the 60S ribosomal subunit and formation of the 80S complex. Abrogation of rRNA methylation did not affect the 48S complex but resulted in impairment of 80S complex assembly on the cellular, but not the viral, IRESs tested. Impairment of 80S complex assembly on the amino acid transporter SNAT2 IRES was rescued by purified 60S subunits containing fully methylated rRNA. We found that rRNA methylation did not affect the activity of any of the viral IRESs tested but affected the activity of numerous cellular IRESs. This work reveals a novel mechanism operating on a cohort of cellular IRESs that involves rRNA methylation for proper 80S complex assembly and efficient translation initiation.
The Drosophila Suppressor of Hairy wing [Su(Hw)] insulator protein has an essential role in the development of the female germline. Here we investigate the function of Su(Hw) in the ovary. We show that Su(Hw) is universally expressed in somatic cells, while germ cell expression is dynamic. Robust levels accumulate in post-mitotic germ cells, where Su(Hw) localization is limited to chromosomes within nurse cells, the specialized cells that support oocyte growth. Although loss of Su(Hw) causes global defects in nurse cell chromosome structure, we demonstrate that these architectural changes are not responsible for the block in oogenesis. Connections between the fertility and insulator functions of Su(Hw) were investigated through studies of the two gypsy insulator proteins, Modifier of (mdg4)67.2 (Mod67.2) and Centrosomal Protein of 190kDa (CP190). Accumulation of these proteins is distinct from Su(Hw), with Mod67.2 and CP190 showing uniform expression in all cells during early stages of oogenesis that diminishes in later stages. Although Mod67.2 and CP190 extensively co-localize with Su(Hw) on nurse cell chromosomes, neither protein is required for nurse cell chromosome development or oocyte production. These data indicate that while the gypsy insulator function requires both Mod67.2 and CP190, these proteins are not essential for oogenesis. These studies represent the first molecular investigations of Su(Hw) function in the germline, which uncover distinct requirements for Su(Hw) insulator and ovary functions.
Jak2/Stat-mediated prolactin signaling culminates in Stat5a-DNA-binding. However, not all Jak2-dependent genes have Stat5 sites. Western analysis with inhibitors showed Jak2 is a proximal intermediate in prolactin-induced RUSH phosphorylation. Transfection assays with HRE-H9 cells showed the RUSH-binding site mediated the ability of prolactin to augment progesterone-dependent transcription of the RUSH gene. Jak2 inhibitors or targeted RUSH-site mutation blocked the prolactin effect. RUSH co-immunoprecipitated with phospho-Jak2 from nuclear extracts. Jak2 inhibitors abolished the nuclear pool of phospho-RUSH not the nuclear content of RUSH in HRE-H9 cells. Nucleolar-affiliated partners, e.g. nucleolin, were identified by microLC/MS/MS analysis of nuclear proteins that co-immunoprecipitated with RUSH/GST-RING. RUSH did not exclusively co-localize with fibrillarin to the nucleolus. MG-132 (proteasomal inhibitor) failed to block Tyrene CR4-mediated decrease in phospho-RUSH, and did not promote RUSH accumulation in the nucleolus. These studies authenticate prolactin-dependent Jak2 phosphorylation of RUSH, and provide functional implications on the RUSH network of nuclear interactions.
The retinoblastoma protein (pRB) negatively regulates cell proliferation by limiting the activity of the family of E2F transcription factors. In Drosophila, mutation of the DEAD-box helicase belle (bel) relieves an E2F/pRB induced G(1) cell cycle arrest; however, the mechanism of this rescue is unknown. Here, we show that the level of the cyclin-dependent kinase inhibitor Dacapo (Dap), homolog of mammalian p21/p27, is strongly reduced both in bel mutant cells in vivo and in tissue culture cells depleted of Bel by RNA interference. Interestingly, the loss of bel also partially alleviates an ectopically induced G(1) cell cycle arrest. Additionally, we show that Bel undergoes nucleocytoplasmic shuttling. Thus, inactivation of bel renders cells less sensitive to several anti-proliferative signals inducing G(1) arrest.
Cellular nutritional and energy status regulates a wide range of nuclear processes important for cell growth, survival, and metabolic homeostasis. Mammalian target of rapamycin (mTOR) plays a key role in the cellular responses to nutrients. However, the nuclear processes governed by mTOR have not been clearly defined. Using isobaric peptide tagging coupled with linear ion trap mass spectrometry, we performed quantitative proteomics analysis to identify nuclear processes in human cells under control of mTOR. Within 3 h of inhibiting mTOR with rapamycin in HeLa cells, we observed down-regulation of nuclear abundance of many proteins involved in translation and RNA modification. Unexpectedly, mTOR inhibition also down-regulated several proteins functioning in chromosomal integrity and up-regulated those involved in DNA damage responses (DDRs) such as 53BP1. Consistent with these proteomic changes and DDR activation, mTOR inhibition enhanced interaction between 53BP1 and p53 and increased phosphorylation of ataxia telangiectasia mutated (ATM) kinase substrates. ATM substrate phosphorylation was also induced by inhibiting protein synthesis and suppressed by inhibiting proteasomal activity, suggesting that mTOR inhibition reduces steady-state (abundance) levels of proteins that function in cellular pathways of DDR activation. Finally, rapamycin-induced changes led to increased survival after radiation exposure in HeLa cells. These findings reveal a novel functional link between mTOR and DDR pathways in the nucleus potentially operating as a survival mechanism against unfavorable growth conditions.
The 35S ribosomal RNA genes (rDNA) are organized as repeated arrays in many organisms. Epigenetic regulation of transcription of the rRNA results in only a subset of copies being transcribed, making rDNA an important model for understanding epigenetic chromatin modification. We have created an allelic series of deletions within the rDNA array of the Drosophila Y chromosome that affect nucleolus size and morphology, but do not limit steady-state rRNA concentrations. These rDNA deletions result in reduced heterochromatin-induced gene silencing elsewhere in the genome, and the extent of the rDNA deletion correlates with the loss of silencing. Consistent with this, chromosomes isolated from strains mutated in genes required for proper heterochromatin formation have very small rDNA arrays, reinforcing the connection between heterochromatin and the rDNA. In wild-type cells, which undergo spontaneous natural rDNA loss, we observed the same correlation between loss of rDNA and loss of heterochromatin-induced silencing, showing that the volatility of rDNA arrays may epigenetically influence gene expression through normal development and differentiation. We propose that the rDNA contributes to a balance between heterochromatin and euchromatin in the nucleus, and alterations in rDNA--induced or natural--affect this balance.
Exposure of cells to DNA-damaging agents results in a rapid increase in the formation of subnuclear complexes containing Rad51. To date, it has not been determined to what extent DNA damage-induced cytoplasmic to nuclear transport of Rad51 may contribute to this process. We have analyzed subcellular fractions of HeLa and HCT116 cells and found a significant increase in nuclear Rad51 levels following exposure to a modest dose of ionizing radiation (2 grays). We also observed a DNA damage-induced increase in nuclear Rad51 in the Brca2-defective cell line Capan-1. To address a possible Brca2-independent mechanism for Rad51 nuclear transport, we analyzed subcellular fractions for two other Rad51-interacting proteins, Rad51C and Xrcc3. Rad51C has a functional nuclear localization signal, and although we found that the subcellular distribution of Xrcc3 was not significantly affected by DNA damage, there was a damage-induced increase in nuclear Rad51C. Furthermore, RNA interference-mediated depletion of Rad51C in HeLa and Capan-1 cells resulted in lower steady-state levels of nuclear Rad51 as well as a diminished DNA damage-induced increase. Our results provide important insight into the cellular regulation of Rad51 nuclear entry and a role for Rad51C in this process.
The ribosomal protein L22 is a component of the 60S eukaryotic ribosomal subunit. As an RNA-binding protein, it has been shown to interact with both cellular and viral RNAs including 28S rRNA and the Epstein-Barr virus encoded RNA, EBER-1. L22 is localized to the cell nucleus where it accumulates in nucleoli. Although previous studies demonstrated that a specific amino acid sequence is required for nucleolar localization, the RNA-binding domain has not been identified. Here, we investigated the hypothesis that the nucleolar accumulation of L22 is linked to its ability to bind RNA. To address this hypothesis, mutated L22 proteins were generated to assess the contribution of specific amino acids to RNA binding and protein localization. Using RNA-protein binding assays, we demonstrate that basic amino acids 80-93 are required for high affinity binding of 28S rRNA and EBER-1 by L22. Fluorescence localization studies using GFP-tagged mutated L22 proteins further reveal that basic amino acids 80-93 are critical for nucleolar accumulation and for incorporation into ribosomes. Our data support the growing consensus that the nucleolar accumulation of ribosomal proteins may not be mediated by a defined localization signal, but rather by specific interaction with established nucleolar components such as rRNA.
CSRP3 or muscle LIM protein (MLP) is a nucleocytoplasmic shuttling protein and a mechanosensor in cardiac myocytes. MLP regulation and function was studied in cultured neonatal rat myocytes treated with pharmacological or mechanical stimuli. Either verapamil or BDM decreased nuclear MLP while phenylephrine and cyclic strain increased it. These results suggest that myocyte contractility regulates MLP subcellular localization. When RNA polymerase II was inhibited with alpha-amanitin, nuclear MLP was reduced by 30%. However, when both RNA polymerase I and II were inhibited with actinomycin D, there was a 90% decrease in nuclear MLP suggesting that its nuclear translocation is regulated by both nuclear and nucleolar transcriptional activity. Using cell permeable synthetic peptides containing the putative nuclear localization signal (NLS) of MLP, nuclear import of the protein in cultured rat neonatal myocytes was inhibited. The NLS of MLP also localizes to the nucleolus. Inhibition of nuclear translocation prevented the increased protein accumulation in response to phenylephrine. Furthermore, cyclic strain of myocytes after prior NLS treatment to remove nuclear MLP resulted in disarrayed sarcomeres. Increased protein synthesis and brain natriuretic peptide expression were also prevented suggesting that MLP is required for remodeling of the myofilaments and gene expression. These findings suggest that nucleocytoplasmic shuttling MLP plays an important role in the regulation of the myocyte remodeling and hypertrophy and is required for adaptation to hypertrophic stimuli.
Retroviruses use different strategies to regulate transcription and translation and exploit the cellular machinery involved in these processes. This study shows that the signal peptide of the envelope glycoprotein (Env) of Jaagsiekte sheep retrovirus (JSRV) plays a major role in posttranscriptional viral gene expression. Expression of the JSRV Env in trans increases viral particle production by mechanisms dependent on (i) its leader sequence, (ii) an intact signal peptide cleavage site, (iii) a cis-acting RNA-responsive element located in the viral genome, (iv) Crm1, and (v) B23. The signal peptide of the JSRV Env (JSE-SP) is 80 amino acid residues in length and contains putative nuclear localization and export signals, in addition to an arginine-rich RNA binding motif. JSE-SP localizes both in the endoplasmic reticulum and in the nucleus, where it colocalizes with nucleolar markers. JSE-SP is a multifunctional protein, as it moderately enhances nuclear export of unspliced viral mRNA and considerably increases viral particle release by favoring a posttranslational step of the replication cycle.
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) is associated with the angioproliferative KS lesions characterized by spindle-shaped endothelial cells, inflammatory cells, cytokines, growth factors, and angiogenic factors. De novo KSHV infection of human microvascular dermal endothelial cells results in increased secretion of several growth factors, cytokines, chemokines, and angiogenic factors, and the multifunctional angiogenic protein angiogenin is one of them. KS tissue sections were positive for angiogenin, highlighting the importance of angiogenin in KS pathogenesis. Examination of KSHV-mediated angiogenin upregulation and secretion and potential outcomes revealed that during infection of primary endothelial cells, KSHV induced a time- and dose-dependent increase in angiogenin gene expression and protein secretion beginning as early as 8 h postinfection and lasting until the fifth day of our observation period. TIVE latently transformed cells (TIVE-LTC) latently infected with KSHV secreted high levels of angiogenin. Angiogenin was also detected in BCBL-1 cells (human B cells) carrying KSHV in a latent state. Significant induction of angiogenin was observed in cells expressing KSHV ORF73 (LANA-1; latent) and ORF74 (lytic) genes alone, and moderate induction was seen with the lytic KSHV ORF50 gene. Angiogenin bound to surface actin, internalized in a microtubule-independent manner, and translocated into the nucleus and nucleolus of infected cells. In addition, it increased 45S rRNA gene transcription, antiapoptosis, and proliferation of infected cells, thus demonstrating the multifunctional nature of KSHV-induced angiogenin. These activities were dependent on angiogenin nuclear translocation, which was inhibited by neomycin. Upregulation of angiogenin led to increased activation of urokinase plasminogen activator and generation of active plasmin, which facilitated the migration of endothelial cells toward chemoattractants, including angiogenin, and chemotaxis was prevented by the inhibition of angiogenin nuclear translocation. Treatment of KSHV-infected cell supernatants with antiangiogenin antibodies significantly inhibited endothelial tube formation, and inhibition of nuclear translocation of angiogenin also blocked the expression of KSHV-induced vascular endothelial growth factor C. Collectively, these results strongly suggest that by increasing infected endothelial cell 45S rRNA synthesis, proliferation, migration, and angiogenesis, KSHV-induced angiogenin could be playing a pivotal role in the pathogenesis of KSHV infection, including a contribution to the angioproliferative nature of KS lesions. Our studies suggested that LANA-1 and vGPCR play roles in KSHV-induced angiogenesis and that the angiogenic potential of vGPCR might also be due to its ability to induce angiogenin.
The thymus is a vertebrate-specific organ where T lymphocytes are generated. Genetic programs that lead to thymus development are incompletely understood. We previously screened ethylnitrosourea-induced medaka mutants for recessive defects in thymus development. Here we report that one of those mutants is caused by a missense mutation in a gene encoding the previously uncharacterized protein WDR55 carrying the tryptophan-aspartate-repeat motif. We find that WDR55 is a novel nucleolar protein involved in the production of ribosomal RNA (rRNA). Defects in WDR55 cause aberrant accumulation of rRNA intermediates and cell cycle arrest. A mutation in WDR55 in zebrafish also leads to analogous defects in thymus development, whereas WDR55-null mice are lethal before implantation. These results indicate that WDR55 is a nuclear modulator of rRNA synthesis, cell cycle progression, and embryonic organogenesis including teleost thymus development.
Drosophila neuroblasts and ovarian stem cells are well characterized models for stem cell biology. In both cell types, one daughter cell self-renews continuously while the other undergoes a limited number of divisions, stops to proliferate mitotically and differentiates. Whereas neuroblasts segregate the Trim-NHL (tripartite motif and Ncl-1, HT2A and Lin-41 domain)-containing protein Brain tumour (Brat) into one of the two daughter cells, ovarian stem cells are regulated by an extracellular signal from the surrounding stem cell niche. After division, one daughter cell looses niche contact. It undergoes 4 transit-amplifying divisions to form a cyst of 16 interconnected cells that reduce their rate of growth and stop to proliferate mitotically. Here we show that the Trim-NHL protein Mei-P26 (refs 7, 8) restricts growth and proliferation in the ovarian stem cell lineage. Mei-P26 expression is low in stem cells but is strongly induced in 16-cell cysts. In mei-P26 mutants, transit-amplifying cells are larger and proliferate indefinitely leading to the formation of an ovarian tumour. Like brat, mei-P26 regulates nucleolar size and can induce differentiation in Drosophila neuroblasts, suggesting that these genes act through the same pathway. We identify Argonaute-1, a component of the RISC complex, as a common binding partner of Brat and Mei-P26, and show that Mei-P26 acts by inhibiting the microRNA pathway. Mei-P26 and Brat have a similar domain composition that is also found in other tumour suppressors and might be a defining property of a new family of microRNA regulators that act specifically in stem cell lineages.
Mutations affecting NPM1 (nucleophosmin) are the most common genetic lesions found in acute myeloid leukemia (AML). NPM1 is one of the most abundant proteins found in the nucleolus and has links to the MDM2/p53 tumor suppressor pathway. A distinctive feature of NPM1 mutants in AML is their aberrant localization to the cytoplasm of leukemic cells. This mutant phenotype is the result of the substitution of several C-terminal residues, including one or two conserved tryptophan residues, with a leucine-rich nuclear export signal. The exact molecular mechanism underlying the loss of nucleolar retention, and the role of the tryptophans, remains unknown. In this study we have determined the structure of an independently folded globular domain in the C terminus of NPM1 using NMR spectroscopy, and we report that the conserved tryptophans are critical for structure. This domain is necessary for the nucleolar targeting of NPM1 and is disrupted by mutations in AML with cytoplasmic NPM1. Furthermore, we identify conserved surface-exposed lysine residues that are functionally rather than structurally important for nucleolar localization. This study provides new focus for efforts to understand the pathogenesis of AML with cytoplasmic NPM1 and may be used to aid the design of small molecules that target the C-terminal domain of NPM1 to act as novel anti-proliferative and anti-leukemia therapeutics.
We have previously shown that the leader proteinase (L(pro)) of foot-and-mouth disease virus (FMDV) interferes with the innate immune response by blocking the translation of interferon (IFN) protein and by reducing the immediate-early induction of beta IFN mRNA and IFN-stimulated genes. Here, we report that L(pro) regulates the activity of nuclear factor kappaB (NF-kappaB). Analysis of NF-kappaB-dependent reporter gene expression in BHK-21 cells demonstrated that infection with wild-type (WT) virus has an inhibitory effect compared to infection with a genetically engineered mutant lacking the leader coding region. The expression of endogenous NF-kappaB-dependent genes tumor necrosis factor alpha and RANTES is also reduced in WT virus-infected primary porcine cells. This inhibitory effect is neither the result of a decrease in the level of the mRNA of p65/RelA, a subunit of NF-kappaB, nor a block on the nuclear translocation of p65/RelA, but instead appears to be a consequence of the degradation of accumulated p65/RelA. Viral L(pro) is localized to the nucleus of infected cells, and there is a correlation between the translocation of L(pro) and the decrease in the amount of nuclear p65/RelA. By using a recombinant cardiovirus expressing L(pro), we demonstrate that the disappearance of p65/RelA takes place in the absence of any other FMDV product. The observation that L(pro) disrupts the integrity of NF-kappaB suggests a global mechanism by which FMDV antagonizes the cellular innate immune and inflammatory responses to viral infection.
Interphase centromeres are crucial domains for the proper assembly of kinetochores at the onset of mitosis. However, it is not known whether the centromere structure is under tight control during interphase. This study uses the peculiar property of the infected cell protein 0 of herpes simplex virus type 1 to induce centromeric structural damage, revealing a novel cell response triggered by centromere destabilization. It involves centromeric accumulation of the Cajal body-associated coilin and fibrillarin as well as the survival motor neuron proteins. The response, which we have termed interphase centromere damage response (iCDR), was observed in all tested human and mouse cells, indicative of a conserved mechanism. Knockdown cells for several constitutive centromere proteins have shown that the loss of centromeric protein B provokes the centromeric accumulation of coilin. We propose that the iCDR is part of a novel safeguard mechanism that is dedicated to maintaining interphase centromeres compatible with the correct assembly of kinetochores, microtubule binding, and completion of mitosis.
Disassembly of the nucleolus during mitosis is driven by phosphorylation of nucleolar proteins. RNA processing stops until completion of nucleolar reformation in G(1) phase. Here, we describe the RNA methyltransferase NSUN2, a novel substrate of Aurora-B that contains an NOL1/NOP2/sun domain. NSUN2 was concentrated in the nucleolus during interphase and was distributed in the perichromosome and cytoplasm during mitosis. Aurora-B phosphorylated NSUN2 at Ser139. Nucleolar proteins NPM1/nucleophosmin/B23 and nucleolin/C23 were associated with NSUN2 during interphase. In mitotic cells, association between NPM1 and NSUN2 was inhibited, but NSUN2-S139A was constitutively associated with NPM1. The Aurora inhibitor Hesperadin induced association of NSUN2 with NPM1 even in mitosis, despite the silver staining nucleolar organizer region disassembly. In vitro methylation experiments revealed that the Aurora-B-phosphorylation and the phosphorylation-mimic mutation (S139E) suppressed methyltransferase activities of NSUN2. These results indicate that Aurora-B participates to regulate the assembly of nucleolar RNA-processing machinery and the RNA methyltransferase activity of NSUN2 via phosphorylation at Ser139 during mitosis.
Hepatitis delta virus (HDV) genome replication requires the virus-encoded small delta protein (deltaAg). During replication, nucleotide sequence changes accumulate on the HDV RNA, leading to the translation of deltaAg species that are nonfunctional or even inhibitory. A replication system was devised where all deltaAg was conditionally provided from a separate and unchanging source. A line of human embryonic kidney cells was stably transfected with a single copy of cDNA encoding small deltaAg, with expression under tetracycline (TET) control. Next, HDV genome replication was initiated in these cells by transfection with a mutated RNA unable to express deltaAg. Thus, replication of this RNA was under control of the TET-inducible deltaAg. In the absence of TET, there was sufficient deltaAg to allow a low level of HDV replication that could be maintained for at least 1 year. When TET was added, both deltaAg and genomic RNA increased dramatically within 2 days. With clones of such cells, designated 293-HDV, the burst of HDV RNA replication interfered with cell cycling. Within 2 days, there was a fivefold enhancement of G1/G0 cells relative to both S and G2/M cells, and by 6 days, there was extensive cell detachment and death. These findings and those of other studies that are under way demonstrate the potential applications of this experimental system.
Nuclease-resistant, cytoplasmically resident molecular beacons were used to specifically label beta-actin mRNA in living and motile chicken embryonic fibroblasts. beta-actin mRNA signals were most abundant in active lamellipodia, which are protrusions that cells extend to adhere to surfaces. Time-lapse images show that the immediate sources of beta-actin mRNA for nascent lamellipodia are adjacent older protrusions. During the development of this method, we observed that conventional molecular beacons are rapidly sequestered in cell nuclei, leaving little time for them to find and bind to their cytoplasmic mRNA targets. By linking molecular beacons to a protein that tends to stay within the cytoplasm, nuclear sequestration was prevented, enabling cytoplasmic mRNAs to be detected and imaged. Probing beta-actin mRNA with these cytoplasmically resident molecular beacons did not affect the motility of the fibroblasts. Furthermore, mRNAs bound to these probes undergo translation within the cell. The use of cytoplasmically resident molecular beacons will enable further studies of the mechanism of beta-actin mRNA localization, and will be useful for understanding the dynamics of mRNA distribution in other living cells.
BACKGROUND:
Inside the cell, the HIV Tat protein is mainly found in the nucleus and nucleolus. The nucleolus, the site of ribosome biogenesis, is a highly organized, non-membrane-bound sub-compartment where proteins with a high affinity for nucleolar components are found. While it is well known that Tat accumulates in the nucleolus via a specific nucleolar targeting sequence, its function in this compartment it still unknown.
RESULTS:
To clarify the significance of the Tat nucleolar localization, we induced the expression of the protein during oogenesis in Drosophila melanogaster strain transgenic for HIV-tat gene. Here we show that Tat localizes in the nucleoli of Drosophila oocyte nurse cells, where it specifically co-localizes with fibrillarin. Tat expression is accompanied by a significant decrease of cytoplasmic ribosomes, which is apparently related to an impairment of ribosomal rRNA precursor processing. Such an event is accounted for by the interaction of Tat with fibrillarin and U3 snoRNA, which are both required for pre-rRNA maturation.
CONCLUSION:
Our data contribute to understanding the function of Tat in the nucleolus, where ribosomal RNA synthesis and cell cycle control take place. The impairment of nucleolar pre-rRNA maturation through the interaction of Tat with fibrillarin-U3snoRNA complex suggests a process by which the virus modulates host response, thus contributing to apoptosis and protein shut-off in HIV-uninfected cells.
Visualization of specific genomic loci in live cells is a prerequisite for the investigation of dynamic changes in chromatin architecture during diverse biological processes, such as cellular aging. However, current precision genomic imaging methods are hampered by the lack of fluorescent probes with high specificity and signal-to-noise contrast. We find that conventional transcription activator-like effectors (TALEs) tend to form protein aggregates, thereby compromising their performance in imaging applications. Through screening, we found that fusing thioredoxin with TALEs prevented aggregate formation, unlocking the full power of TALE-based genomic imaging. Using thioredoxin-fused TALEs (TTALEs), we achieved high-quality imaging at various genomic loci and observed aging-associated (epi) genomic alterations at telomeres and centromeres in human and mouse premature aging models. Importantly, we identified attrition of ribosomal DNA repeats as a molecular marker for human aging. Our study establishes a simple and robust imaging method for precisely monitoring chromatin dynamics in vitro and in vivo.
Endonuclease VIII-like protein 1 (NEIL1) is a DNA glycosylase involved in initiating the base excision repair pathway, the major cellular mechanism for repairing DNA base damage. Here, we have purified the major E3 ubiquitin ligases from human cells responsible for regulation of NEIL1 by ubiquitylation. Interestingly, we have identified two enzymes that catalyse NEIL1 polyubiquitylation, Mcl-1 ubiquitin ligase E3 (Mule) and tripartite motif 26 (TRIM26). We demonstrate that these enzymes are capable of polyubiquitylating NEIL1 in vitro, and that both catalyse ubiquitylation of NEIL1 within the same C-terminal lysine residues. An siRNA-mediated knockdown of Mule or TRIM26 leads to stabilisation of NEIL1, demonstrating that these enzymes are important in regulating cellular NEIL1 steady state protein levels. Similarly, a mutant NEIL1 protein lacking residues for ubiquitylation is more stable than the wild type protein in vivo We also demonstrate that cellular NEIL1 protein is induced in response to ionising radiation (IR), although this occurs specifically in a Mule-dependent manner. Finally we show that stabilisation of NEIL1, particularly following TRIM26 siRNA, contributes to cellular resistance to IR. This highlights the importance of Mule and TRIM26 in maintaining steady state levels of NEIL1, but also those required for the cellular DNA damage response.
The histone deacetylase (HDAC) inhibitor vorinostat has received significant attention in recent years as an 'epigenetic' drug used to treat solid tumors. However, its mechanisms of action are not entirely understood, particularly with regard to its interaction with the aberrations in 3D nuclear structure that accompany neoplastic progression. We investigated the impact of vorinostat on human esophageal epithelial cell lines derived from normal, metaplastic (pre-cancerous), and malignant tissue. Using a combination of novel optical computed tomography (CT)-based quantitative 3D absorption microscopy and conventional confocal fluorescence microscopy, we show that subjecting malignant cells to vorinostat preferentially alters their 3D nuclear architecture relative to non-cancerous cells. Optical CT (cell CT) imaging of fixed single cells showed that drug-treated cancer cells exhibit significant alterations in nuclear morphometry. Confocal microscopy revealed that vorinostat caused changes in the distribution of H3K9ac-marked euchromatin and H3K9me3-marked constitutive heterochromatin. Additionally, 3D immuno-FISH showed that drug-induced expression of the DNA repair gene MGMT was accompanied by spatial relocation toward the center of the nucleus in the nuclei of metaplastic but not in non-neoplastic cells. Our data suggest that vorinostat's differential modulation of 3D nuclear architecture in normal and abnormal cells could play a functional role in its anti-cancer action.
The yeast 2-micron plasmid epitomizes the evolutionary optimization of selfish extra-chromosomal genomes for stable persistence without jeopardizing their hosts' fitness. Analyses of fluorescence-tagged single-copy reporter plasmids and/or the plasmid partitioning proteins in native and non-native hosts reveal chromosome-hitchhiking as the likely means for plasmid segregation. The contribution of the partitioning system to equal segregation is bipartite- replication-independent and replication-dependent. The former nearly eliminates 'mother bias' (preferential plasmid retention in the mother cell) according to binomial distribution, thus limiting equal segregation of a plasmid pair to 50%. The latter enhances equal segregation of plasmid sisters beyond this level, elevating the plasmid close to chromosome status. Host factors involved in plasmid partitioning can be functionally separated by their participation in the replication-independent and/or replication-dependent steps. In the hitchhiking model, random tethering of a pair of plasmids to chromosomes signifies the replication-independent component of segregation; the symmetric tethering of plasmid sisters to sister chromatids embodies the replication-dependent component. The 2-micron circle broadly resembles the episomes of certain mammalian viruses in its chromosome-associated propagation. This unifying feature among otherwise widely differing selfish genomes suggests their evolutionary convergence to the common logic of exploiting, albeit via distinct molecular mechanisms, host chromosome segregation machineries for self-preservation.
MAP1LC3B (microtubule-associated protein 1 light chain 3, LC3) is a key component of the autophagy pathway, contributing to both cargo selection and autophagosome formation in the cytoplasm. Emerging evidence suggests that nuclear forms of LC3 are also functionally important; however, the mechanisms that facilitate the nuclear targeting and trafficking of LC3 between the nucleus and cytoplasm under steady-state conditions are poorly understood. In this study, we examine how residues known to regulate the interactions between LC3 and other proteins or RNA (F52 L53, R68-R70 and G120) contribute to its nuclear targeting, nucleocytoplasmic transport and association with nucleoli and other nuclear components. We find that residues F52 L53 and R68-70, but not G120, regulate targeting of LC3 to the nucleus, its rates of nucleocytoplasmic transport and the apparent sizes of LC3-associated complexes in the nucleus inferred from fluorescence recovery after photobleaching (FRAP) measurements. We also show that LC3 is enriched in nucleoli and its triple arginine motif is especially important for nucleolar targeting. Finally, we identify a series of candidate nuclear LC3-interacting proteins using mass spectrometry, including MAP1B, tubulin and several 40S ribosomal proteins. These findings suggest LC3 is retained in the nucleus in association with high-molecular weight complexes that continuously scan the nucleolus.
Melanoma antigen D2 (MAGE-D2) is recognized as a cancer diagnostic marker; however, it has poorly characterized functions. Here, we established its intracellular localization and shuttling during cell cycle progression and in response to cellular stress. In normal conditions, MAGE-D2 is present in the cytoplasm, nucleoplasm, and nucleoli. Within the latter, MAGE-D2 is mostly found in the granular and the dense fibrillar components, and it interacts with nucleolin. Transfection of MAGE-D2 deletion mutants demonstrated that Δ203-254 leads to confinement of MAGE-D2 to the cytoplasm, while Δ248-254 prevents its accumulation in nucleoli but still allows its presence in the nucleoplasm. Consequently, this short sequence belongs to a nucleolar localization signal. MAGE-D2 deletion does not alter the nucleolar organization or rRNA levels. However, its intracellular localization varies with the cell cycle in a different kinetic than nucleolin. After genotoxic and nucleolar stresses, MAGE-D2 is excluded from nucleoli and concentrates in the nucleoplasm. We demonstrated that its camptothecin-related delocalization results from two distinct events: a rapid nucleolar release and a slower phospho-ERK-dependent cytoplasm to nucleoplasm translocation, which results from an increased flux from the cytoplasm to nucleoplasm. In conclusion, MAGE-D2 is a dynamic protein whose shuttling properties could suggest a role in cell cycle regulation.
Mutations in ANKRD11 have recently been reported to cause KBG syndrome, an autosomal dominant condition characterized by intellectual disability (ID), behavioral problems, and macrodontia. To understand the pathogenic mechanism that relates ANKRD11 mutations with the phenotype of KBG syndrome, we studied the cellular characteristics of wild-type ANKRD11 and the effects of mutations in humans and mice. We show that the abundance of wild-type ANKRD11 is tightly regulated during the cell cycle, and that the ANKRD11 C-terminus is required for the degradation of the protein. Analysis of 11 pathogenic ANKRD11 variants in humans, including six reported in this study, and one reported in the Ankrd11 (Yod/+) mouse, shows that all mutations affect the C-terminal regions and that the mutant proteins accumulate aberrantly. In silico analysis shows the presence of D-box sequences that are signals for proteasome degradation. We suggest that ANKRD11 C-terminus plays an important role in regulating the abundance of the protein, and a disturbance of the protein abundance due to the mutations leads to KBG syndrome.
Nesprins are a multi-isomeric family of spectrin-repeat (SR) proteins, predominantly known as nuclear envelope scaffolds. However, isoforms that function beyond the nuclear envelope remain poorly examined. Here, we characterize p50(Nesp1), a 50-kD isoform that localizes to processing bodies (PBs), where it acts as a microtubule-associated protein capable of linking mRNP complexes to microtubules. Overexpression of dominant-negative p50(Nesp1) caused Rck/p54, but not GW182, displacement from microtubules, resulting in reduced PB movement and cross talk with stress granules (SGs). These cells disassembled canonical SGs induced by sodium arsenite, but not those induced by hydrogen peroxide, leading to cell death and revealing PB-microtubule attachment is required for hydrogen peroxide-induced SG anti-apoptotic functions. Furthermore, p50(Nesp1) was required for miRNA-mediated silencing and interacted with core miRISC silencers Ago2 and Rck/p54 in an RNA-dependent manner and with GW182 in a microtubule-dependent manner. These data identify p50(Nesp1) as a multi-functional PB component and microtubule scaffold necessary for RNA granule dynamics and provides evidence for PB and SG micro-heterogeneity.
Kaposi's sarcoma-associated herpesvirus (KSHV) is an oncogenic herpesvirus associated with multiple AIDS-related malignancies. Like other herpesviruses, KSHV has a biphasic life cycle and both the lytic and latent phases are required for tumorigenesis. Evidence suggests that KSHV lytic replication can cause genome instability in KSHV-infected cells, although no mechanism has thus far been described. A surprising link has recently been suggested between mRNA export, genome instability and cancer development. Notably, aberrations in the cellular transcription and export complex (hTREX) proteins have been identified in high-grade tumours and these defects contribute to genome instability. We have previously shown that the lytically expressed KSHV ORF57 protein interacts with the complete hTREX complex; therefore, we investigated the possible intriguing link between ORF57, hTREX and KSHV-induced genome instability. Herein, we show that lytically active KSHV infected cells induce a DNA damage response and, importantly, we demonstrate directly that this is due to DNA strand breaks. Furthermore, we show that sequestration of the hTREX complex by the KSHV ORF57 protein leads to this double strand break response and significant DNA damage. Moreover, we describe a novel mechanism showing that the genetic instability observed is a consequence of R-loop formation. Importantly, the link between hTREX sequestration and DNA damage may be a common feature in herpesvirus infection, as a similar phenotype was observed with the herpes simplex virus 1 (HSV-1) ICP27 protein. Our data provide a model of R-loop induced DNA damage in KSHV infected cells and describes a novel system for studying genome instability caused by aberrant hTREX.
Dosage compensation in Drosophila involves a global activation of genes on the male X chromosome. The activating complex (MSL-DCC) consists of male-specific-lethal (MSL) proteins and two long, noncoding roX RNAs. The roX RNAs are essential for X-chromosomal targeting, but their contributions to MSL-DCC structure and function are enigmatic. Conceivably, the RNA helicase MLE, itself an MSL subunit, is actively involved in incorporating roX into functional DCC. We determined the secondary structure of roX2 and mapped specific interaction sites for MLE in vitro. Upon addition of ATP, MLE disrupted a functionally important stem loop in roX2. This RNA remodeling enhanced specific ATP-dependent association of MSL2, the core subunit of the MSL-DCC, providing a link between roX and MSL subunits. Probing the conformation of roX in vivo revealed a remodeled stem loop in chromatin-bound roX2. The active remodeling of a stable secondary structure by MLE may constitute a rate-limiting step for MSL-DCC assembly.
RNA polymerase I (Pol I) transcription is essential for the cell cycle, growth and protein synthesis in eukaryotes. In the present study, we found that phosphatidylinositol 4,5-bisphosphate (PIP2) is a part of the protein complex on the active ribosomal promoter during transcription. PIP2 makes a complex with Pol I and the Pol I transcription factor UBF in the nucleolus. PIP2 depletion reduces Pol I transcription, which can be rescued by the addition of exogenous PIP2. In addition, PIP2 also binds directly to the pre-rRNA processing factor fibrillarin (Fib), and co-localizes with nascent transcripts in the nucleolus. PIP2 binding to UBF and Fib modulates their binding to DNA and RNA, respectively. In conclusion, PIP2 interacts with a subset of Pol I transcription machinery, and promotes Pol I transcription.
In this study we report that expression of glioma pathogenesis-related protein 1 (GLIPR1) regulated numerous apoptotic, cell cycle, and spindle/centrosome assembly-related genes, including AURKA and TPX2, and induced apoptosis and/or mitotic catastrophe (MC) in prostate cancer (PCa) cells, including p53-mutated/deleted, androgen-insensitive metastatic PCa cells. Mechanistically, GLIPR1 interacts with heat shock cognate protein 70 (Hsc70); this interaction is associated with SP1 and c-Myb destabilization and suppression of SP1- and c-Myb-mediated AURKA and TPX2 transcription. Inhibition of AURKA and TPX2 using siRNA mimicked enforced GLIPR1 expression in the induction of apoptosis and MC. Recombinant GLIPR1-ΔTM protein inhibited AURKA and TPX2 expression, induced apoptosis and MC, and suppressed orthotopic xenograft tumor growth. Our results define a novel GLIPR1-regulated signaling pathway that controls apoptosis and/or mitotic catastrophe in PCa cells and establishes the potential of this pathway for targeted therapies.
In Drosophila melanogaster, as in many animal and plant species, centromere identity is specified epigenetically. In proliferating cells, a centromere-specific histone H3 variant (CenH3), named Cid in Drosophila and Cenp-A in humans, is a crucial component of the epigenetic centromere mark. Hence, maintenance of the amount and chromosomal location of CenH3 during mitotic proliferation is important. Interestingly, CenH3 may have different roles during meiosis and the onset of embryogenesis. In gametes of Caenorhabditis elegans, and possibly in plants, centromere marking is independent of CenH3. Moreover, male gamete differentiation in animals often includes global nucleosome for protamine exchange that potentially could remove CenH3 nucleosomes. Here we demonstrate that the control of Cid loading during male meiosis is distinct from the regulation observed during the mitotic cycles of early embryogenesis. But Cid is present in mature sperm. After strong Cid depletion in sperm, paternal centromeres fail to integrate into the gonomeric spindle of the first mitosis, resulting in gynogenetic haploid embryos. Furthermore, after moderate depletion, paternal centromeres are unable to re-acquire normal Cid levels in the next generation. We conclude that Cid in sperm is an essential component of the epigenetic centromere mark on paternal chromosomes and it exerts quantitative control over centromeric Cid levels throughout development. Hence, the amount of Cid that is loaded during each cell cycle appears to be determined primarily by the preexisting centromeric Cid, with little flexibility for compensation of accidental losses.
The neurodegenerative disorder ataxia with oculomotor apraxia 2 (AOA-2) is caused by defects in senataxin, a putative RNA/DNA helicase thought to be involved in the termination of transcription at RNA polymerase pause sites. RNA/DNA hybrids (R loops) that arise during transcription pausing lead to genome instability unless they are resolved efficiently. We found that senataxin forms distinct nuclear foci in S/G(2)-phase human cells and that the number of these foci increases in response to impaired DNA replication or DNA damage. Senataxin colocalizes with 53BP1, a key DNA damage response protein, and with other factors involved in DNA repair. Inhibition of transcription using α-amanitin, or the dissolution of R loops by transient expression of RNase H1, leads to the loss of senataxin foci. These results indicate that senataxin localizes to sites of collision between components of the replisome and the transcription apparatus and that it is targeted to R loops, where it plays an important role at the interface of transcription and the DNA damage response.
Raptor is the key scaffolding protein that recruits mTOR substrates to rapamycin-sensitive mTOR complex 1 (mTORC1), a molecular integrator of mitogenic and nutrient/energy environmental inputs into protein translation and cell growth. Although Raptor phosphorylation on various sites is pivotal in the regulation of mTORC1 activity, it remains to be elucidated whether site-specific phosphorylation differentially distributes Raptor to unique subcellular compartments. When exploring the spatiotemporal cell cycle dynamics of six different phospho (P)-Raptor isoforms (Thr ( 706) , Ser ( 722) , Ser ( 863) , Ser ( 792) and Ser ( 877) ), a number of remarkable events differentially defined a topological resetting of P-RaptorThr706 on interphasic and mitotic chromosomes. In interphase nuclei, P-Raptor (Thr706) co-localized with fibrillarin, a component of the nucleolar small nuclear ribonucleoprotein particle, as well as with RNA polymerase I, the enzyme that transcribes nucleolar rRNA. Upon Actinomycin D-induced nucleolar segregation and disaggregation, P-RaptorThr706 was excluded from the nucleolus to accumulate at discrete nucleoplasmic bodies. During mitosis, CDK1 inhibition-induced premature assembly of nucleoli relocated fibrillarin to the surrounding regions of chromosomal-associated P-Raptor (Thr706) , suggesting that a subpopulation of mitotic P-Raptor (Thr706) remained targeted at chromosomal loops of rDNA or nuclear organizer regions (NORs). At the end of mitosis and cytokinesis, when reassembly of incipient nucleoli begins upon NORs activation of rDNA transcription, fibrillarin spatially reorganized with P-Raptor (Thr706) to give rise to daughter nucleoli. Treatment with IGF1 exclusively hyperactivated nuclear P-Raptor (Ser706) and concomitantly promoted Ser ( 2481) autophosphorylation of mTOR, which monitors mTORC1-associated catalytic activity. Nucleolar- and NOR-associated P-Raptor (Ser706) may physically link mTORC1 signaling to ever-growing nucleolus plurifunctionality including ribosome biogenesis, cell stress sensor and cell cycle/aging control.
γ-Tubulin is assumed to be a typical cytosolic protein necessary for nucleation of microtubules from microtubule organizing centers. Using immunolocalization and cell fractionation techniques in combination with siRNAi and expression of FLAG-tagged constructs, we have obtained evidence that γ-tubulin is also present in nucleoli of mammalian interphase cells of diverse cellular origins. Immunoelectron microscopy has revealed γ-tubulin localization outside fibrillar centers where transcription of ribosomal DNA takes place. γ-Tubulin was associated with nucleolar remnants after nuclear envelope breakdown and could be translocated to nucleoli during mitosis. Pretreatment of cells with leptomycin B did not affect the distribution of nuclear γ-tubulin, making it unlikely that rapid active transport via nuclear pores participates in the transport of γ-tubulin into the nucleus. This finding was confirmed by heterokaryon assay and time-lapse imaging of photoconvertible protein Dendra2 tagged to γ-tubulin. Immunoprecipitation from nuclear extracts combined with mass spectrometry revealed an association of γ-tubulin with tumor suppressor protein C53 located at multiple subcellular compartments including nucleoli. The notion of an interaction between γ-tubulin and C53 was corroborated by pull-down and co-immunoprecipitation experiments. Overexpression of γ-tubulin antagonized the inhibitory effect of C53 on DNA damage G(2) /M checkpoint activation. The combined results indicate that aside from its known role in microtubule nucleation, γ-tubulin may also have nuclear-specific function(s).
How steroid hormones shape animal growth remains poorly understood. In Drosophila, the main steroid hormone, ecdysone, limits systemic growth during juvenile development. Here we show that ecdysone controls animal growth rate by specifically acting on the fat body, an organ that retains endocrine and storage functions of the vertebrate liver and fat. We demonstrate that fat body-targeted loss of function of the Ecdysone receptor (EcR) increases dMyc expression and its cellular functions such as ribosome biogenesis. Moreover, changing dMyc levels in this tissue is sufficient to affect animal growth rate. Finally, the growth increase induced by silencing EcR in the fat body is suppressed by cosilencing dMyc. In conclusion, the present work reveals an unexpected function of dMyc in the systemic control of growth in response to steroid hormone signaling.
Dimethylation of histone H3 lysine 9 (H3K9me2) is an important epigenetic mark associated with transcription repression. Here, we identified PHF8, a JmjC-domain-containing protein, as a histone demethylase specific for this repressing mark. Recombinant full-length wild type protein could remove methylation from H3K9me2, but mutation of a conserved histidine to alanine H247A abolished the demethylase activity. Overexpressed exogenous PHF8 was colocalized with B23 staining. Endogenous PHF8 was also colocalized with B23 and fibrillarin, two well-established nucleolus proteins, suggesting that PHF8 is localized in the nucleolus and may regulate rRNA transcription. Indeed, PHF8 bound to the promoter region of the rDNA gene. Knockdown of PHF8 reduced the expression of rRNA, and overexpression of the gene resulted in upregulation of rRNA transcript. Concomitantly, H3K9me2 level was elevated in the promoter region of the rDNA gene in PHF8 knockdown cells and reduced significantly when the wild type but not the catalytically inactive H247A mutant PHF8 was overexpressed. Thus, our study identified a histone demethylase for H3K9me2 that regulates rRNA transcription.
In female marsupials, X chromosome inactivation (XCI) is imprinted, affecting the paternal X chromosome. One model, supported by recent studies, proposes that XCI in marsupials is achieved through inheritance of an already silent X chromosome from the father, with XCI initiated by meiotic sex chromosome inactivation (MSCI). This model is appealing because marsupials have no Xist gene and the marsupial inactive X chromosome is epigenetically dissimilar to that of mice, apparently lacking repressive histone marks such as H3K27 trimethylation. A central prediction of the meiotic inactivation model of XCI is that silencing of genes on the X chromosome, initiated during male meiosis, is stably maintained during subsequent spermiogenesis. Here we characterize XCI in the male germline and female soma of the marsupial Monodelphis domestica. Contrary to the meiotic inactivation model, we find that X genes silenced by MSCI are reactivated after meiosis and are subsequently inactivated in the female. A reexamination of the female somatic inactive marsupial X chromosome reveals that it does share common properties with that of eutherians, including H3K27 trimethylation and targeting to the perinucleolar compartment. We conclude that aspects of the XCI process are more highly conserved in therian mammals than previously thought.
Structure-specific endonucleases resolve DNA secondary structures generated during DNA repair and recombination. The yeast 5' flap endonuclease Slx1-Slx4 has received particular attention with the finding that Slx4 has Slx1-independent key functions in genome maintenance. Although Slx1 is a highly conserved protein in eukaryotes, no orthologs of Slx4 were reported other than in fungi. Here we report the identification of Slx4 orthologs in metazoa, including fly MUS312, essential for meiotic recombination, and human BTBD12, an ATM/ATR checkpoint kinase substrate. Human SLX1-SLX4 displays robust Holliday junction resolvase activity in addition to 5' flap endonuclease activity. Depletion of SLX1 and SLX4 results in 53BP1 foci accumulation and H2AX phosphorylation as well as cellular hypersensitivity to MMS. Furthermore, we show that SLX4 binds the XPF(ERCC4) and MUS81 subunits of the XPF-ERCC1 and MUS81-EME1 endonucleases and is required for DNA interstrand crosslink repair. We propose that SLX4 acts as a docking platform for multiple structure-specific endonucleases.
Growth and body size are regulated by the CNS, integrating the genetic developmental program with assessments of an animal's current energy state and environmental conditions. CNS decisions are transmitted to all cells of the animal by insulin/insulin-like signals. The molecular biology of the CNS growth control system has remained, for the most part, elusive. Here we identify NS3, a Drosophila nucleostemin family GTPase, as a powerful regulator of body size. ns3 mutants reach <60% of normal size and have fewer and smaller cells, but exhibit normal body proportions. NS3 does not act cell-autonomously, but instead acts at a distance to control growth. Rescue experiments were performed by expressing wild-type ns3 in many different cells of ns3 mutants. Restoring NS3 to only 106 serotonergic neurons rescued global growth defects. These neurons are closely apposed with those of insulin-producing neurons, suggesting possible communication between the two neuronal systems. In the brains of ns3 mutants, excess serotonin and insulin accumulate, while peripheral insulin pathway activation is low. Peripheral insulin pathway activation rescues the growth defects of ns3 mutants. The findings suggest that NS3 acts in serotonergic neurons to regulate insulin signaling and thus exert global growth control.
The use of cardenolides like ouabain, digitoxin, or oleandrin has been reported previously many times as a means of potentially combating human refractory prostate cancer by inducing apoptosis through an increase in intracellular calcium concentrations. The aims of the current study were to investigate if part of the antitumor effects mediated by cardenolides concerned disorganization of nucleolar structure and whether this was further associated with a marked decrease in c-Myc expression. Accordingly, the antitumor activity of a novel hemisynthetic cardenolide [1R,3aS,3bR,5aS,6aR,7aS,9R,12aR,13aR,15aR]-3a,11a-dihydroxy-13a-(hydroxymethyl)-9,15a-dimethyl-1-(5-oxo-2,5-dihydrofuran-3-yl)icosahydro-1H,4'H-spiro[cyclopenta [7,8]phenanthro[2,3-b]pyrano[3,2-e][1,4]dioxine-11,2'-[1,3]thiazolidin]-4'-one (UNBS1450)] was compared with that of classic cardenolides and reference anticancer agents in prostate cancer cell lines in vitro and in vivo following s.c. and orthotopic prostate cancer cell grafting into mice. The present study indicates that UNBS1450 markedly decreases the in vitro viability/proliferation of human prostate cancer cell lines but not of normal cells. The induced effects are not linked to an increase in intracellular calcium concentrations and subsequent induction of apoptosis. Rather, they appear to relate to the compound's capacity to disorganize nucleolar structure and function (through an impairment of cyclin-dependent kinase and c-Myc expression and related signaling pathways; paralleled by the disorganization of cancer cell-specific perinucleolar bodies as revealed by disruption of Sam68). This nonapoptotic cancer cell death mediated by severe nucleolar targeting and down-regulation of c-Myc expression is a completely new cardenolide-induced mechanism of antitumor action.
Pluripotent embryonic stem (ES) cells must select between alternative fates of self-replication and lineage commitment during continuous proliferation. Here, we delineate the role of autocrine production of fibroblast growth factor 4 (Fgf4) and associated activation of the Erk1/2 (Mapk3/1) signalling cascade. Fgf4 is the major stimulus activating Erk in mouse ES cells. Interference with FGF or Erk activity using chemical inhibitors or genetic ablations does not impede propagation of undifferentiated ES cells. Instead, such manipulations restrict the ability of ES cells to commit to differentiation. ES cells lacking Fgf4 or treated with FGF receptor inhibitors resist neural and mesodermal induction, and are refractory to BMP-induced non-neural differentiation. Lineage commitment potential of Fgf4-null cells is restored by provision of FGF protein. Thus, FGF enables rather than antagonises the differentiation activity of BMP. The key downstream role of Erk signalling is revealed by examination of Erk2-null ES cells, which fail to undergo either neural or mesodermal differentiation in adherent culture, and retain expression of pluripotency markers Oct4, Nanog and Rex1. These findings establish that Fgf4 stimulation of Erk1/2 is an autoinductive stimulus for naïve ES cells to exit the self-renewal programme. We propose that the Erk cascade directs transition to a state that is responsive to inductive cues for germ layer segregation. Consideration of Erk signalling as a primary trigger that potentiates lineage commitment provides a context for reconciling disparate views on the contribution of FGF and BMP pathways during germ layer specification in vertebrate embryos.
Recent transcriptome analyses have revealed that a large body of noncoding regions of mammalian genomes are actually transcribed into RNAs. Our understanding of the molecular features of these noncoding RNAs is far from complete. We have identified a novel mRNA-like noncoding gene, named Gomafu, which is expressed in a distinct set of neurons in the mouse nervous system. Interestingly, spliced mature Gomafu RNA is localized to the nucleus despite its mRNA-like characteristics, which usually act as potent export signals to the cytoplasm. Within the nucleus, Gomafu RNA is detected as numerous spots that do not colocalize with known nuclear domain markers. Gomafu RNA is extremely insoluble and remains intact after nuclear matrix preparation. Furthermore, heterokaryon assays revealed that Gomafu RNA does not shuttle between the nucleus and cytoplasm, but is retained in the nucleus after its transcription. We propose that Gomafu RNA represents a novel family of mRNA-like noncoding RNA that constitutes a cell-type-specific component of the nuclear matrix.
In mammalian females, two X chromosomes are epigenetically distinguished as active and inactive chromosomes to balance X-linked gene dosages between males and females. How the Xs are maintained differently in the same nucleus remains unknown. Here, we demonstrate that the inactive X (Xi) is targeted to a distinct nuclear compartment following pairing with its homologous partner. During mid-to-late S phase, 80%-90% of Xi contact the nucleolus and reside within a Snf2h-enriched ring. Autosomes carrying ectopic X-inactivation center sequences are also targeted to the perinucleolar compartment. Deleting Xist results in a loss of nucleolar association and an inability to maintain Xi heterochromatin, leading to Xi reactivation at the single gene level. We propose that the Xi must continuously visit the perinucleolar compartment to maintain its epigenetic state. These data raise a mechanism by which chromatin states can be replicated by spatial and temporal separation in the nucleus.
How stem cells generate both differentiating and self-renewing daughter cells is unclear. Here, we show that Drosophila larval neuroblasts-stem cell-like precursors of the adult brain-regulate proliferation by segregating the growth inhibitor Brat and the transcription factor Prospero into only one daughter cell. Like Prospero, Brat binds and cosegregates with the adaptor protein Miranda. In larval neuroblasts, both Brat and Prospero are required to inhibit self-renewal in one of the two daughter cells. While Prospero regulates cell-cycle gene transcription, Brat acts as a posttranscriptional inhibitor of dMyc. In brat or prospero mutants, both daughter cells grow and behave like neuroblasts leading to the formation of larval brain tumors. Similar defects are seen in lethal giant larvae (lgl) mutants where Brat and Prospero are not asymmetric. We have identified a molecular mechanism that may control self-renewal and prevent tumor formation in other stem cells as well.
Although hsp70 antagonizes apoptosis-inducing factor (AIF)-mediated cell death, the relative importance of preventing its release from mitochondria versus sequestering leaked AIF in the cytosol remains controversial. To dissect these two protective mechanisms, hsp70 deletion mutants lacking either the chaperone function (hsp70-deltaEEVD) or ATPase function (hsp70-deltaATPase) were selectively overexpressed before exposing cells to a metabolic inhibitor, an insult sufficient to cause mitochondrial AIF release, nuclear AIF accumulation, and apoptosis. Compared with empty vector, overexpression of wild type human hsp70 inhibited bax activation and reduced mitochondrial AIF release after injury. In contrast, mutants lacking either the chaperone function (hsp70-deltaEEVD) or the ATP hydrolytic domain (hsp70-deltaATPase) failed to prevent mitochondrial AIF release. Although hsp70-deltaEEVD did not inhibit bax activation or mitochondrial membrane injury after cell stress, this hsp70 mutant co-immunoprecipitated with leaked AIF in injured cells and decreased nuclear AIF accumulation. In contrast, hsp70-deltaATPase did not interact with AIF either in intact cells or in a cell-free system and furthermore, failed to prevent nuclear AIF accumulation. These results demonstrate that mitochondrial protection against bax-mediated injury requires both intact chaperone and ATPase functions, whereas the ATPase domain is critical for sequestering AIF in the cytosol.
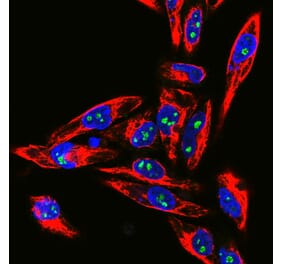
![Western Blot - Anti-Fibrillarin Antibody [ARC0506] (A306505) - Antibodies.com](https://cdn.antibodies.com/image/catalog/306/A306505_1.jpg?profile=product_alternative)



![Western Blot - Anti-Fibrillarin Antibody [ARC0506] (A306505) - Antibodies.com](https://cdn.antibodies.com/image/catalog/306/A306505_1.jpg?profile=product_alternative)
![Immunofluorescence - Anti-Fibrillarin Antibody [38F3] (A85370) - Antibodies.com](https://cdn.antibodies.com/image/catalog/85/A85370_1.jpg?profile=product_top)
![Western Blot - Anti-Fibrillarin Antibody [38F3] (A85370) - Antibodies.com](https://cdn.antibodies.com/image/catalog/85/A85370_2.jpg?profile=product_top)
![Immunofluorescence - Anti-Fibrillarin Antibody [38F3] (A85370) - Antibodies.com](https://cdn.antibodies.com/image/catalog/85/A85370_1.jpg?profile=product_top_thumb)
![Western Blot - Anti-Fibrillarin Antibody [38F3] (A85370) - Antibodies.com](https://cdn.antibodies.com/image/catalog/85/A85370_2.jpg?profile=product_top_thumb)
![Immunofluorescence - Anti-Fibrillarin Antibody [38F3] (A85370) - Antibodies.com](https://cdn.antibodies.com/image/catalog/85/A85370_1.jpg?profile=product_image)
![Western Blot - Anti-Fibrillarin Antibody [38F3] (A85370) - Antibodies.com](https://cdn.antibodies.com/image/catalog/85/A85370_2.jpg?profile=product_image)